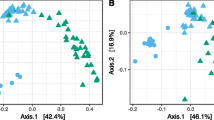
Abstract
Endophytic fungal communities have been shown to be highly diverse in almost every host plant species analyzed so far. However, the factors shaping their compositions are largely unknown. To elucidate the impact of various factors, 10 independent replicates of DNA extracts from each of 17 surface-sterilized leaf and stem samples were analyzed by pyrosequencing of fungal ITS1 rRNA gene amplicons. Altogether, 154 fungal OTUs (operational taxonomic units), represented by 953,385 sequences, were found in at least 2 samples from Viscum album ssp. austriacum and/or its host Pinus sylvestris. Deviating from earlier, cultivation-based assessments, the communities were dominated by OTUs related to the genus Mortierella and OTUs not assignable to a certain fungal phylum. However, Ascomycota were still the most diverse group in terms of OTU richness and already hypothesized organ and host preferences of certain endophytic Xylariaceae isolated from the Pinus-Viscum-system could be confirmed. Host species and organ type were also the major factors shaping the detected fungal communities. The two plant species clearly differed according to the endophytic fungal communities, but only stems and needles of Pinus were inhabited by significantly different fungal assemblages. Interestingly, only the 1 and 3 year old stem sections differed according to the endophytic fungal community, while differently aged leaves of both plants were indistinguishable in this regard. Size of the organs had no impact on fungal communities in Pinus, but shorter internodes and smaller leaves showed at least a tendency to differ from the corresponding larger organs in Viscum. Fungal communities also differed slightly between the two sampling sites, lying 200 km apart, and between the three sampling campaigns. Because the samples were drawn within 15 days, this finding indicates that seasonal shifts clearly outweigh aging effects in host plant with perennial leaves. The results therefore provide strong evidence against a linear development of the endophytic fungal communities in Pinus sylvestris and Viscum album over the years. The communities seem to establish themselves already in the year the respective organs emerge. Further study is required to clarify whether they predominantly establish anew each year, or if the core community persists throughout subsequent years. The most abundant endophytic OTUs are known from soil and/or dead plant material and are supposed to represent latent decomposers. The study reveals for the first time that host and/or organ preferences of putatively saprotrophic fungi are predominantly responsible for compositional differences in the endophytic fungal communities between host plants and organs. While the analyses are shown to provide rather robust results, the significance of genetic abundance, as revealed by high-throughput sequencing analyses, remains an unsettled issue. This is discussed in detail, as well as the challenges in assigning taxonomic names to OTUs.



Similar content being viewed by others
References
Ainsworth AM (1995) Technical information sheet no.11: isolation techniques for basidiomycetes. World J Microbiol Biotechnol 11:364–366. doi:10.1007/bf00367125
Amend AS, Seifert KA, Bruns TD (2010) Quantifying microbial communities with 454 pyrosequencing: does read abundance count? Mol Ecol 19:5555–5565. doi:10.1111/j.1365-294X.2010.04898.x
Arnold AE (2007) Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol Rev 21:51–66
Arnold AE, Mejia LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, Herre EA (2003) Fungal endophytes limit pathogen damage in a tropical tree. PNAS 100:15649–15654
Baldrian P, Větrovský T, Cajthaml T, Dobiášová P, Petránková M, Šnajdr J, Eichlerova I (2013) Estimation of fungal biomass in forest litter and soil. Fungal Ecol 6:1–11. doi:10.1016/j.funeco.2012.10.002
Begerow D, Nilsson H, Unterseher M, Maier W (2010) Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl Microbiol Biotechnol 87:99–108. doi:10.1007/s00253-010-2585-4
Bergsten J, Bilton DT, Fujisawa T, Elliott M, Monaghan MT, Balke M, Hendrich L, Geijer J, Herrmann J, Foster GN, Ribera I, Nilsson AN, Barraclough TG, Vogler AP (2012) The effect of geographical scale of sampling on DNA barcoding. Syst Biol. doi:10.1093/sysbio/sys037
Bissegger M, Sieber TN (1994) Assemblages of endophytic fungi in coppice shoots of Castanea sativa. Mycologia 86:648–655. doi:10.2307/3760535
Bridge PD, Roberts PJ, Spooner BM, Panchal G (2003) On the unreliability of published DNA sequences. New Phytol 160:43–48. doi:10.1046/j.1469-8137.2003.00861.x
Carreiro MM, Koske RE (1992) Room-temperature isolations can bias against selection of low- temperature microfungi in temperate forest soils. Mycologia 84:886–900. doi:10.2307/3760287
Chase MW, Fay MF (2009) Barcoding of plants and fungi. Science 325(5941):682–683. doi:10.1126/science.1176906
Collado J, Platas G, Gonzalez I, Pelaez F (1999) Geographical and seasonal influences on the distribution of fungal endophytes in Quercus ilex. New Phytol 144:525–532
Collado J, Platas G, Paulus B, Bills GF (2007) High-throughput culturing of fungi from plant litter by a dilution-to-extinction technique. FEMS Microbiol Ecol 60:521–533. doi:10.1111/j.1574-6941.2007.00294.x
Davey ML, Heegaard E, Halvorsen R, Kauserud H, Ohlson M (2013) Amplicon-pyrosequencing-based detection of compositional shifts in bryophyte-associated fungal communities along an elevation gradient. Mol Ecol 22:368–383. doi:10.1111/mec.12122
Domsch KH, Gams W, Anderson TH (2007) Compendium of soil fungi. IHW-Verlag, Eching
Fisher PJ, Petrini O, Sutton BC (1993) A comparative study of fungal endophytes in leaves, xylem and bark of Eucalyptus in Australia and England. Sydowia 45:338–345
Fisher PJ, Petrini O, Petrini LE, Sutton BC (1994) Fungal endophytes from the leaves and twigs of Quercus ilex L. from England, Majorca and Switzerland. New Phytol 127:133–137. doi:10.1111/j.1469-8137.1994.tb04267.x
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes–application to the identification of mycorrhizae and rusts. Mol Ecol 2(2):113–118. doi:10.1111/j.1365-294X.1993.tb00005.x
Giordano L, Gonthier P, Varese GC, Miserere L, Nicolotti G (2009) Mycobiota inhabiting sapwood of healthy and declining Scots pine (Pinus sylvestris L.) trees in the Alps. Fungal Divers 38:69–83
Hamilton C, Gundel PE, Helander M, Saikkonen K (2012) Endophytic mediation of reactive oxygen species and antioxidant activity in plants: a review. Fungal Divers 54:1–10. doi:10.1007/s13225-012-0158-9
Hata K, Atari R, Sone K (2002) Isolation of endophytic fungi from leaves of Pasania edulis and their within-leaf distributions. Mycoscience 43:369–373
Hawksworth DL (2012) Global species numbers of fungi: are tropical studies and molecular approaches contributing to a more robust estimate? Biodivers Conserv 21:2425–2433. doi:10.1007/s10531-012-0335-x
Hoffman MT, Arnold AE (2008) Geographic locality and host identity shape fungal endophyte communities in cupressaceous trees. Mycol Res 112:331–344. doi:10.1016/j.mycres.2007.10.014
Johnston PR, Sutherland PW, Joshee S (2006) Visualising endophytic fungi within leaves by detection of (1→3)-ß-d-glucans in fungal cell walls. Mycologist 20:159–162. doi:10.1016/j.mycol.2006.10.003
Jumpponen A, Jones KL (2010) Seasonally dynamic fungal communities in the Quercus macrocarpa phyllosphere differ between urban and nonurban environments. New Phytol 186:496–513. doi:10.1111/j.1469-8137.2010.03197.x
Kowalski T (1993) Fungi in living symptomless needles of Pinus sylvestris with respect to some observed disease processes. J Phytopathol–Phytopathol Z 139:129–145
Kumar DSS, Hyde KD (2004) Biodiversity and tissue-recurrence of endophytic fungi in Tripterygium wilfordii. Fungal Divers 17:69–90
Lindahl BD, Boberg J (2007) Distribution and function of litter basidiomycetes in coniferous forests. In: Boddy L, Frankland JF, van West P (eds) Ecology of saprotrophic basidiomycetes. Elsevier, Amsterdam, pp 183–196
Lodge DJ, Fisher PJ, Sutton BC (1996) Endophytic fungi of Manilkara bidentata leaves in Puerto Rico. Mycologia 88:733–738. doi:10.2307/3760967
Magurran AE, Henderson PA (2003) Explaining the excess of rare species in natural species abundance distributions. Nature 422(6933):714–716
Masarovlčová E (1991) Leaf shape, stomata density and photosynthetic rate of the common oak leaves. Biol Plant 33:495–500. doi:10.1007/bf02897727
Mishra A, Gond S, Kumar A, Sharma V, Verma S, Kharwar R, Sieber T (2012) Season and tissue type affect fungal endophyte communities of the indian medicinal plant Tinospora cordifolia more strongly than geographic location. Microb Ecol 64:388–398. doi:10.1007/s00248-012-0029-7
Nilsson RH, Ryberg M, Kristiansson E, Abarenkov K, Larsson K-H, Koljalg U (2006) Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS One 1(1):e59. doi:10.1371/journal.pone.0000059
Nilsson R, Bok G, Ryberg M, Kristiansson E, Hallenberg N (2009) A software pipeline for processing and identification of fungal ITS sequences. Source Code Biol Med 4:1–6. doi:10.1186/1751-0473-4-1
Osono T (2006) Role of phyllosphere fungi of forest trees in the development of decomposer fungal communities and decomposition processes of leaf litter. Can J Microbiol 52:701–716. doi:10.1139/w06-023
Osono T (2008) Endophytic and epiphytic phyllosphere fungi of Camellia japonica: seasonal and leaf age-dependent variations. Mycologia 100:387–391
Pääkkönen E, Vahala J, Pohjola M, Holopainen T, Kärenlampi L (1998) Physiological, stomatal and ultrastructural ozone responses in birch (Betula pendula Roth.) are modified by water stress. Plant Cell Environ 21:671–684. doi:10.1046/j.1365-3040.1998.00303.x
Pedros-Alio C (2006) Marine microbial diversity: can it be determined? Trends Microbiol 14:257–263. doi:10.1016/j.tim.2006.04.007
Pedrós-Alió C (2012) The rare bacterial biosphere. Annu Rev Mar Sci 4:449–466. doi:10.1146/annurev-marine-120710-100948
Peršoh D, Rambold G (2012) Lichen-associated fungi of the Letharietum vulpinae. Mycol Prog 11:753–760. doi:10.1007/s11557-011-0786-6
Peršoh D, Melcher M, Graf K, Fournier J, Stadler M, Rambold G (2009) Molecular and morphological evidence for the delimitation of Xylaria hypoxylon. Mycologia 101(2):256–268. doi:10.3852/08-108
Peršoh D, Melcher M, Flessa F, Rambold G (2010) First fungal community analyses of endophytic ascomycetes associated with Viscum album ssp. austriacum and its host Pinus sylvestris. Fungal Biol 114:585–596. doi:10.1016/j.funbio.2010.04.009
Peršoh D, Weig AR, Rambold G (2012) A transcriptome—targeting EcoChip for assessing functional mycodiversity. Microarrays 1:25–41. doi:10.3390/microarrays1010025
Peršoh D, Segert J, Zigan A, Rambold G (2013) Fungal community composition shifts along a leaf degradation gradient in a European beech forest. Plant Soil 362:175–186. doi:10.1007/s11104-012-1271-y
Petkovits T, Nagy LG, Hoffmann K, Wagner L, Nyilasi I, Griebel T, Schnabelrauch D, Vogel H, Voigt K, Vágvölgyi C, Papp T (2011) Data partitions, bayesian analysis and phylogeny of the Zygomycetous fungal family Mortierellaceae, inferred from nuclear ribosomal DNA sequences. PLoS One 6:e27507. doi:10.1371/journal.pone.0027507
Petrini O, Fisher PJ (1990) Occurence of fungal endophytes in twigs of Salix fragilis and Quercus robur. Mycol Res 94:1077–1080
Porras-Alfaro A, Bayman P (2011) Hidden fungi, emergent properties: endophytes and microbiomes. Annu Rev Phytopathol 49:291–315. doi:10.1146/annurev-phyto-080508-081831
Promputtha I, Lumyong S, Dhanasekaran V, McKenzie EH, Hyde KD, Jeewon R (2007) A phylogenetic evaluation of whether endophytes become saprotrophs at host senescence. Microb Ecol 53:579–590. doi:10.1007/s00248-006-9117-x
Purahong W, Hyde K (2011) Effects of fungal endophytes on grass and non-grass litter decomposition rates. Fungal Divers 47:1–7. doi:10.1007/s13225-010-0083-8
R Development Core Team (2008) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org
Ragazzi A, Morrica S, Capretti P, Dellavalle I, Mancini F, Turco E (2001) Endophytic fungi in Quercus cerris: isolation frequency in relation to phenological phase, tree health and the organ affected. Phytopathol Mediterr 40:165–171
Ragazzi A, Moricca S, Capretti P, Dellavalle I, Turco E (2003) Differences in composition of endophytic mycobiota in twigs and leaves of healthy and declining Quercus species in Italy. Forest Pathol 33:31–38
Raidl S, Bonfigli R, Agerer R (2005) Calibration of quantitative real-time TaqMan PCR by correlation with hyphal biomass and ITS copies in mycelia of Piloderma croceum. Plant Biol 7:713–717. doi:10.1055/s-2005-873003
Rodriguez RJ, White JF, Arnold AE, Redman RS (2009) Fungal endophytes: diversity and functional roles. New Phytol 182:314–330
Santamaria O, Diez JJ (2005) Fungi in leaves, twigs and stem bark of Populus tremula from northern Spain. For Pathol 35(2):95–104
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding, Consortium (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. PNAS 109:6241–6246. doi:10.1073/pnas.1117018109
Scholtysik A, Unterseher M, Otto P, Wirth C (2012) Spatio-temporal dynamics of endophyte diversity in the canopy of European ash (Fraxinus excelsior). Mycol Prog, in press. doi:10.1007/s11557-012-0835-9
Sieber TN (2007) Endophytic fungi in forest trees: are they mutualists? Fungal Biol Rev 21:75–89
Sieber TN, Siebercanavesi F, Dorworth CE (1991) Endophytic fungi of Red Alder (Alnus rubra) leaves and twigs in British-Columbia. Can J Bot-Rev Can Bot 69:407–411
Sun X, Ding Q, Hyde KD, Guo LD (2012) Community structure and preference of endophytic fungi of three woody plants in a mixed forest. Fungal Ecol 5:624–632. doi:10.1016/j.funeco.2012.04.001
Suryanarayanan T (2011) Diversity of fungal endophytes in tropical trees. In: Pirttilä AM, Frank AC (eds) Endophytes of forest trees, vol 80. Forestry sciences. Springer, Netherlands, pp 67–80. doi:10.1007/978-94-007-1599-8_4
Tellenbach C, Gruenig CR, Sieber TN (2010) Suitability of quantitative real-time PCR to estimate the biomass of fungal root endophytes. Appl Environ Microbiol 76:5764–5772. doi:10.1128/aem.00907-10
Thorn RG, Reddy CA, Harris D, Paul EA (1996) Isolation of saprophytic basidiomycetes from soil. Appl Environ Microbiol 62:4288–4292
Unterseher M (2011) Diversity of fungal endophytes in temperate forest trees. In: Pirttilä AM, Frank AC (eds) Endophytes of forest trees, vol 80. Forestry sciences. Springer, Netherlands, pp 31–46. doi:10.1007/978-94-007-1599-8_2
Unterseher M, Schnittler M (2009) Dilution-to-extinction cultivation of leaf-inhabiting endophytic fungi in beech (Fagus sylvatica L.)–different cultivation techniques influence fungal biodiversity assessment. Mycol Res 113:645–654. doi:10.1016/j.mycres.2009.02.002
Unterseher M, Reiher A, Finstermeier K, Otto P, Morawetz W (2007) Species richness and distribution patterns of leaf-inhabiting endophytic fungi in a temperate forest canopy. Mycol Prog 6:201–212. doi:10.1007/s11557-007-0541-1
Unterseher M, Jumpponen ARI, ÖPik M, Tedersoo L, Moora M, Dormann CF, Schnittler M (2011) Species abundance distributions and richness estimations in fungal metagenomics–lessons learned from community ecology. Mol Ecol 20:275–285. doi:10.1111/j.1365-294X.2010.04948.x
Unterseher M, Peršoh D, Schnittler M (2013) Leaf-inhabiting endophytic fungi of European Beech (Fagus sylvatica L.) co-occur in leaf litter but are rare on decaying wood of the same host. Fungal Divers. doi:10.1007/s13225-013-0222-0
Wearn JA, Sutton BC, Morley NJ, Gange AC (2012) Species and organ specificity of fungal endophytes in herbaceous grassland plants. J Ecol 100:1085–1092. doi:10.1111/j.1365-2745.2012.01997.x
Weig A, Peršoh D, Werner S, Betzlbacher A, Rambold G (2013) Diagnostic assessment of mycodiversity in environmental samples by fungal ITS1 rDNA length polymorphism. Mycol Prog. doi:10.1007/s11557-012-0883-1
Weiß M, Sýkorová Z, Garnica S, Riess K, Martos F, Krause C, Oberwinkler F, Bauer R, Redecker D (2011) Sebacinales everywhere: previously overlooked ubiquitous fungal endophytes. PLoS One 6:e16793. doi:10.1371/journal.pone.0016793
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand D, Shinsky J, White T (eds) PCR protocols: a guide to methods and applications. Academic Press, pp 315–322
Zamora P, Martinez-Ruiz C, Diez JJ (2008) Fungi in needles and twigs of pine plantations from northern Spain. Fungal Divers 30:171–184
Zhou D, Hyde K (2001) Host-specificity, host-exclusivity, and host-recurrence in saprobic fungi. Mycol Res 105:1449–1457
Acknowledgments
I thank Reinhard Agerer (Munich) and Gerhard Rambold (Bayreuth) for fruitful discussions and access to their laboratories, as well as Anina Neumann and Sofia Tello (both Munich) for technical assistance. The study was funded by the Deutsche Forschungsgemeinschaft (DFG, project PE 1673/2).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 43 kb)
Tab. S1
Details on the assignment of contigs. The assigned OTU for each contig is given in the second column. The most similar sequence found in GenBank is listed in the following section with the corresponding Bit-Score. The assigned name is listed in the third part, together with the “Bit Score” of the worst matching sequence considered for the name assignment. The sequences considered for name assignment (i.e. sequences obtaining “Bit Scores” which are at least 0.9 times as high as the “Bit Score” the best matching sequence obtained), are categorized as matches (i.e. sequences deposited under names matching the assigned name), ambiguities (i.e. sequences deposited under names neither confirming nor objecting the assigned name), and the number of outliers (i.e. sequences deposited under names not considered for the name assignment). The number of sequences from uncultured organisms among those considered for name assignment is given in the following column. The final section list the extracted ITS1 sequence of each contig and its length. (XLSX 42 kb)
Tab. S2
Absolute abundance of contigs in samples. The sampling categories for each sample are given in columns 163–171. (XLSX 108 kb)
Fig. S1
Average abundance of fungal taxa. The relative abundance of sequences is summarized for each family. Higher taxonomic affiliations were chosen in case of uncertain family assignment of the corresponding OTUs. Bars for the unclassifiable Mycota (Mycota inc. sed.: 47 % of all sequences) and the Mortierellaceae (33 %) are truncated. (PPTX 65 kb)
Rights and permissions
About this article
Cite this article
Peršoh, D. Factors shaping community structure of endophytic fungi–evidence from the Pinus-Viscum-system. Fungal Diversity 60, 55–69 (2013). https://doi.org/10.1007/s13225-013-0225-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-013-0225-x