Abstract
Purpose
Prevalence of organic pollutants or their natural analogs in soil is often assumed to lead to adaptation in the bacterial community, which results in enhanced bioremediation if the soil is later contaminated. In this study, the effects of soil type and contamination history on diesel oil degradation and bacterial adaptation were studied.
Methods
Mesocosms of mineral and organic forest soil (humus) were artificially treated with diesel oil, and oil hydrocarbon concentrations (GC-FID), bacterial community composition (denaturing gradient gel electrophoresis, DGGE), and oil hydrocarbon degraders (DGGE + sequencing of 16S rRNA genes) were monitored for 20 weeks at 16°C.
Results
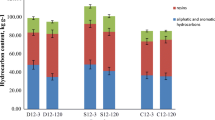
Degradation was advanced in previously contaminated soils as compared with pristine soils and in coniferous organic forest soil as compared with mineral soil. Contamination affected bacterial community composition especially in the pristine mineral soil, where diesel addition increased the number of strong bands in the DGGE gel. Sequencing of cloned 16S rRNA gene fragments and DGGE bands showed that potential oil-degrading bacteria were found in mineral and organic soils and in both pristine and previously contaminated mesocosms. Fast oil degradation was not associated with the presence of any particular bacterial strain in soil.
Conclusions
We demonstrate at the mesocosm scale that previously contaminated and coniferous organic soils are superior environments for fast oil degradation as compared with pristine and mineral soil environments. These results may be utilized in preventing soil pollution and planning soil remediation.




Similar content being viewed by others
References
Aislabie J, McLeod M, Fraser R (1998) Potential for biodegradation of hydrocarbons in soil from the Ross Dependency, Antarctica. Appl Microbiol Biotechnol 49:210–214
Aislabie J, Balks M, Astori N, Stevenson G, Symons R (1999) Polycyclic aromatic hydrocarbons in fuel-oil contaminated soils, Antarctica. Chemosphere 39:2201–2207
Aislabie J, Foght J, Saul D (2000) Aromatic hydrocarbon-degrading bacteria from soil near Scott Base, Antarctica. Polar Biol 23:183–188
Aislabie J, Saul DJ, Foght JM (2006) Bioremediation of hydrocarbon-contaminated polar soils. Extremophiles 10:171–179
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL (2000) GenBank. Nucleic Acids Res 28:15–18
Bundy JG, Paton GI, Campbell CD (2002) Microbial communities in different soil types do not converge after diesel contamination. J Appl Microbiol 92:276–288
Chaineau CH, Morel J, Dupont J, Bury E, Oudot J (1999) Comparison of the fuel oil biodegradation potential of hydrocarbon-assimilating microorganisms isolated from a temperate agricultural soil. Sci Total Environ 227:237–247
Gentry TJ, Josephson KL, Pepper IL (2004) Functional establishment of introduced chlorobenzoate degraders following bioaugmentation with newly activated soil—enhanced contaminant remediation via activated soil bioaugmentation. Biodegradation 15:67–75
Greenwood PF, Wibrow S, George SJ, Tibbett M (2009) Hydrocarbon biodegradation and soil microbial community response to repeated oil exposure. Org Geochem 40:293–300
Hultman J, Pitkäranta M, Romantschuk M, Auvinen P, Paulin L (2008) Probe-based negative selection for underrepresented phylotypes in large environmental clone libraries. J Microbiol Methods 75:457–463
Kanerva S, Kitunen V, Kiikkilä O, Loponen J, Smolander A (2006) Response of soil C and N transformations to tannin fractions originating from Scots pine and Norway spruce needles. Soil Biol Biochem 38:1364–1374
Kanerva T, Palojärvi A, Rämö K, Manninen S (2008) Changes in soil microbial community structure under elevated tropospheric O3 and CO2. Soil Biol Biochem 40:2502–2510
Kauppi S, Sinkkonen A, Romantschuk M (2011) Enhancing bioremediation of diesel-fuel-contaminated soil in a boreal climate: comparison of biostimulation and bioaugmentation. Int Biodeterior Biodegrad 65:359–368
Koivula TT, Salkinoja-Salonen M, Peltola R, Romantschuk M (2004) Pyrene degradation in forest humus microcosms with or without pine and its mycorrhizal fungus. J Environ Qual 33:45–53
Kurola J, Salkinoja-Salonen M, Aarnio T, Hultman J, Romantschuk M (2005) Activity, diversity and population size of ammonia-oxidising bacteria in oil-contaminated landfarming soil. FEMS Microbiol Lett 250:33–38
Labbe D, Margesin R, Schinner F, Whyte LG, Greer CW (2007) Comparative phylogenetic analysis of microbial communities in pristine and hydrocarbon-contaminated Alpine soils. FEMS Microbiol Ecol 59:466–475
Labud V, Garcia C, Hernandez T (2007) Effect of hydrocarbon pollution on the microbial properties of a sandy and a clay soil. Chemosphere 66:1863–1871
Leahy JG, Colwell RR (1990) Microbial-degradation of hydrocarbons in the environment. Microbiol Rev 54:305–315
Margesin R (2000) Potential of cold-adapted microorganisms for bioremediation of oil-polluted Alpine soils. Int Biodeterior Biodegrad 46:3–10
Margesin R, Schinner F (1999a) Biodegradation of diesel oil by cold-adapted microorganisms in presence of sodium dodecyl sulfate. Chemosphere 38:3463–3472
Margesin R, Schinner F (1999b) Biological decontamination of oil spills in cold environments. J Chem Technol Biotechnol 74:381–389
Margesin R, Haemmerle M, Tscherko D (2007) Microbial activity and community composition during bioremediation of diesel-oil-contaminated soil: effects of hydrocarbon concentration, fertilizers, and incubation time. Microb Ecol 53:259–269
Muyzer G (1999) DGGE/TGGE a method for identifying genes from natural ecosystems. Curr Opin Microbiol 2:317–322
Muyzer G, Smalla K (1998) Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127–141
Muyzer G, Dewaal EC, Uitterlinden AG (1993) Profiling of complex microbial-populations by denaturing gradient gel-electrophoresis analysis of polymerase chain reaction-amplified genes-coding for 16s ribosomal-RNA. Appl Environ Microbiol 59:695–700
Peltola R, Salkinoja-Salonen M, Pulkkinen J, Koivunen M, Turpeinen AR, Aarnio T, Romantschuk M (2006) Nitrification in polluted soil fertilized with fast- and slow-releasing nitrogen: a case study at a refinery landfarming site. Environ Pollut 143:247–253
Penet S, Vendeuvre C, Bertoncini F, Marchal R, Monot F (2006) Characterisation of biodegradation capacities of environmental microflorae for diesel oil by comprehensive two-dimensional gas chromatography. Biodegradation 17:577–585
Romantschuk M, Sarand I, Petänen T, Peltola R, Jonsson-Vihanne M, Koivula T, Yrjälä K, Haahtela K (2000) Means to improve the effect of in situ bioremediation of contaminated soil: an overview of novel approaches. Environ Pollut 107:179–185
Sarand I, Haario H, Jorgensen KS, Romantschuk M (2000) Effect of inoculation of a TOL plasmid containing mycorrhizosphere bacterium on development of Scots pine seedlings, their mycorrhizosphere and the microbial flora in m-toluate-amended soil. FEMS Microbiol Ecol 31:127–141
Sarand I, Skarfstad E, Forsman M, Romantschuk M, Shingler V (2001) Role of the DmpR-mediated regulatory circuit in bacterial biodegradation properties in methylphenol-amended soils. Appl Environ Microbiol 67:162–171
Saul DJ, Aislabie JM, Brown CE, Harris L, Foght JM (2005) Hydrocarbon contamination changes the bacterial diversity of soil from around Scott Base, Antarctica. FEMS Microbiol Ecol 53:141–155
Schlueter PM, Harris SA (2006) Analysis of multilocus fingerprinting data sets containing missing data. Mol Ecol Notes 6:569–572
Scragg A (2005) Environmental biotechnology. Oxford University Press, Oxford
Sjøholm OR, Aamand J, Sørensen J, Nybroe O (2010) Degrader density determines spatial variability of 2,6-dichlorobenzamide mineralisation in soil. Environ Pollut 158:292–298
Springael D, Top EM (2004) Horizontal gene transfer and microbial adaptation to xenobiotics: new types of mobile genetic elements and lessons from ecological studies. Trends Microbiol 12:53–58
Suni S, Malinen E, Kosonen J, Silvennoinen H, Romantschuk M (2007) Electrokinetically enhanced bioremediation of creosote-contaminated soil: laboratory and field studies. J Environ Sci Health Part A–Toxic/Hazard Subst Environ Eng 42:277–287
Tan X, Chang SX (2007) Soil compaction and forest litter amendment affect carbon and net nitrogen mineralization in a boreal forest soil. Soil Tillage Res 93:77–86
Timmis KN (ed) (2010) Handbook of hydrocarbon and lipid microbiology. Springer, Berlin
Vogel TM (1996) Bioaugmentation as a soil bioremediation approach. Curr Opin Biotechnol 7:311–316
Welander U (2005) Microbial degradation of organic pollutants in soil in a cold climate. Soil Sediment Contam 14:281–291
Wheeler DL, Chappey C, Lash AE, Leipe DD, Madden TL, Schuler GD, Tatusova TA, Rapp BA (2000) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 28:10–14
Widmer F, Seidler RJ, Gillevet PM, Watrud LS, Di Giovanni GD (1998) A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Appl Environ Microbiol 64:2545–2553
Yang S-Z, Jin H-J, Wei Z, He R-X, Ji Y-J, Li X-M, Yu S-P (2009) Bioremediation of oil spills in cold environments: a review. Pedosphere 19:371–381
Acknowledgments
The study was funded by Foundation for Research of Natural Resources in Finland, Marjatta and Eino Kolli Foundation and Elite project funded by ERDF and the Regional Council of Päijät-Häme. The authors wish to thank Hanna Puttonen, Mari Joensuu, and Tomasz Płociniczak for the laboratory assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Zhihong Xu
Rights and permissions
About this article
Cite this article
Kauppi, S., Romantschuk, M., Strömmer, R. et al. Natural attenuation is enhanced in previously contaminated and coniferous forest soils. Environ Sci Pollut Res 19, 53–63 (2012). https://doi.org/10.1007/s11356-011-0528-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-011-0528-y