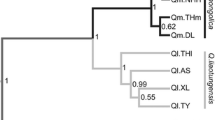
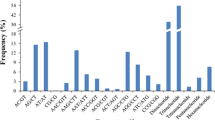
Abstract
Oaks (Quercus: Fagaceae) are ecological and economic keystones of many forested ecosystems but effective genetic management strategies are hindered by high levels of phenotypic plasticity within species and frequent hybridization among them. These same features, however, make oak communities suited for the study of speciation, hybridization, and genetic adaptation. Efforts to develop new and to adapt existing genomic resources to less-studied members of this genus should not only improve oak conservation and management but also aid the study of fundamental evolutionary processes. Here, we present a suite of 27 highly polymorphic simple sequence repeat (SSR) markers tested in four North American red oak (Quercus section Lobatae) species: Q. rubra, Q. ellipsoidalis, Q. coccinea, and Q. velutina. Five markers are genomic SSRs (gSSRs) — four novel and one previously transferred from Q. petraea — and 22 are gene-based SSRs derived from Q. robur and Q. petraea expressed sequence tags (EST-SSRs). Overall, levels of polymorphism detected with these primer pairs were high, with gene diversity (H e) averaging 0.66 across all loci in natural populations. In addition, we show that EST-SSR markers may have the potential to detect divergent selection at stress-resistance candidate genes among closely related oak species.
Similar content being viewed by others
Notes
Sequence data have been deposited in the GenBank, DDBJ Nucleotide Sequence Database and EMBL databases under the accession numbers JQ929659 (locus 1P10), JQ929660 (2P24), JQ929661 (3A05), and JQ929662 (3D15).
References
Abrams MD (1988) Comparative water relationships of three successional hardwood species in central Wisconsin. Tree Physiol 4:263–273. doi:10.1093/treephys/4.3.263
Abrams MD (1990) Adaptations and responses to drought in Quercus species of North America. Tree Physiol 7:227–238. doi:10.1093/treephys/7.1-2-3-4.227
Aldrich PR, Cavender-Bares J (2011) Quercus. In: Kole C (ed) Wild crop relatives: Genomic and breeding resources, forest trees. Springer, Berlin, pp 89–129, doi: 10.1007/978-3-642-21250-5
Aldrich PR, Glaubitz JC, Parker GR, Rhodes OE, Michler CH (2005) Genetic structure inside a declining red oak community in old-growth forest. J Hered 96:627–634. doi:10.1093/jhered/esi115
Aldrich PR, Parker GR, Michler CH, Romero-Severson J (2003a) Whole-tree silvic identifications and the microsatellite genetic structure of a red oak species complex in an Indiana old-growth forest. Can J For Res 33:2228–2237. doi:10.1139/x03-160
Aldrich PR, Jagtap M, Michler CH, Romero-Severson J (2003b) Amplification of North American red oak microsatellite markers in European white oaks and Chinese chestnut. Silvae Genet 52:176–179
Aldrich PR, Michler CH, Sun WL, Romero-Severson J (2002) Microsatellite markers for northern red oak (Fagaceae: Quercus rubra). Mol Ecol Notes 2:472–474. doi:10.1046/j.1471-8278.2002.00282.x
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi:10.1093/nar/25.17.3389
Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G (2008) LOSITAN: a workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinforma 8:323. doi:10.1186/1471-2105-9-323
Barreneche T, Casasoli M, Russell K, Akkak A, Meddour H, Plomion C, Villani F, Kremer A (2004) Comparative mapping between Quercus and Castanea using simple-sequence repeats (SSRs). Theor Appl Genet 108:558–566
Beaumont MA (2005) Adaptation and speciation: what can Fst tell us? Trends Ecol Evol 20:435–440
Beaumont MA, Nichols RA (1996) Evaluating loci for use in the genetic analysis of population structure. Proc Roc Soc B 263:1619–1626
Blue MP, Jensen RJ (1988) Positional and season variaion in oak (Quercus: Fagaceae) leaf morphology. Am J Bot 75:939–947. doi:10.2307/2443759
Bodénès C, Joandet S, Laigret F, Kremer A (1997) Detection of genomic regions differentiating two closely related oak species Quercus petraea (Matt.) Liebl. and Quercus robur L. Heredity 78:433–444. doi:10.1038/hdy.1997.67
Bouck A, Vision T (2007) The molecular ecologist's guide to expressed sequence tags. Mol Ecol 16:907–924. doi:10.1111/j.1365-294X.2006.03195.x
Bruschi P, Vendramin GG, Bussotti F, Grossoni P (2000) Morphological and molecular differentiation between Quercus petraea (Matt.) Liebl. and Quercus pubescens Willd. (Fagaceae) in Northern and Central Italy. Ann Bot 85:325–333. doi:10.1006/anbo.1999.1046
Castillo A, Dorado G, Feuillet C, Sourdille P, Hernandez P (2010) Genetic structure and ecogeographical adaptation in wild barley (Hordeum chilense Roemer et Schultes) as revealed by microsatellite markers. BMC Plant Biol 10. doi:10.1186/1471-2229-10-266
Charlesworth D (2006) Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet 2(4): e64. doi:10.1371/journal.pgen.0020064
Clotfelter ED, Pedersen AB, Cranford JA, Ram N, Snajdr EA, Nolan V, Ketterson ED (2007) Acorn mast drives long-term dynamics of rodent and songbird populations. Oecologia 154:493–503. doi:10.1007/s00442-007-0859-z
Coart E, Lamote V, De Loose M, Van Bockstaele E, Lootens P, Roldan-Ruiz I (2002) AFLP markers demonstrate local genetic differentiation between two indigenous oak species Quercus robur L. and Quercus petraea (Matt.) Liebl in Flemish populations. Theor Appl Genet 105:431–439. doi:10.1007/s00122-002-0920-6
Curtu AL, Gailing O, Finkeldey R (2007a) Evidence for hybridization and introgression within a species-rich oak (Quercus spp.) community. BMC Evol Biol 7:218. doi:10.1186/1471-2148-7-218
Curtu AL, Gailing O, Leinemann L, Finkeldey R (2007b) Genetic variation and differentiation within a natural community of five oak species (Quercus spp.). Plant Biol 9:116–126. doi:10.1055/s-2006-924542
Durand J, Bodénès C, Chancerel E, Frigero J-M, Vendramin GG, Sebastiani F, Buonamici A, Gailing O, Koelewijn H-P, Villani F, Mattioni C, Cherubini M, Goicoechea PG, Herran A, Ikaran Z, Cabane C, Ueno S, de Daruvar A, Kremer A, Plomion C (2010) A fast and cost-effective approach to develop and map EST-SSR markers: oak as a case study. BMC Genomics 11:570. doi:10.1186/1471-2164-11-570
Ellis JR, Burke JM (2007) EST-SSRs as a resource for population genetic analyses. Heredity 99:125–132. doi:10.1038/sj.hdy.6801001
Fernández J, Sork V, Gallego G, López J, Bohorques A, Tohme J (2000) Cross-amplification of microsatellite loci in a neotropical Quercus species and standardization of DNA extraction from mature leaves dried in silica gel. Plant Mol Biol Rep 18:397–397. doi:10.1007/bf02825070
Forkner RE, Hunter MD (2000) What goes up must come down? Nutrient addition and predation pressure on oak herbivores. Ecology 81:1588–1600. doi:10.2307/177309
Gerwein JB, Kesseli RV (2006) Genetic diversity and population structure of Quercus rubra (Fagaceae) in old-growth and secondary forests in southern New England. Rhodora 108:1–18. doi:10.3119/05-9.1
Guo Y, Huang C, Xie Y, Song F, Zhou X (2010) A tomato glutaredoxin gene SIGRX1 regulates plant responses to oxidative, drought and salt stresses. Planta 232:1499–1509. doi:10.1007/s00425-010-1271-1
Guttman SI, Weigt LA (1989) Electrophoretic evidence of relationships among Quercus (oaks) of eastern North America. Can J Bot 67:339–351
Hipp AL, Weber JA (2008) Taxonomy of Hill's oak (Quercus ellipsoidalis: Fagaceae): evidence from AFLP data. Syst Bot 33:148–158. doi:10.1600/036364408783887320
Hokanson SC, Isebrands JG, Jensen RJ, Hancock JF (1993) Isozyme variation in oaks of the Apostle Islands in Wisconsin — genetic structure and levels of inbreeding in Quercus rubra and Q. ellipsoidalis (Fagaceae). Am J Bot 80:1349–1357. doi:10.2307/2445720
Kalinowski ST (2004) Counting alleles with rarefaction: private alleles and hierarchical sampling designs. Conserv Genet 5:539–543. doi:10.1023/B:COGE.0000041021.91777.1a
Kalinowski ST (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189. doi:10.1111/j.1471-8286.2004.00845.x
Kremer A, Kleinschmit J, Cottrell J, Cundall EP, Deans JD, Ducousso A, Konig AO, Lowe AJ, Munro RC, Petit RJ, Stephan BR (2002) Is there a correlation between chloroplastic and nuclear divergence, or what are the roles of history and selection on genetic diversity in European oaks? For Ecol Manage 156:75–87. doi:10.1016/s0378-1127(01)00635-1
Liewlaksaneeyanawin C, Ritland CE, El-Kassaby YA, Ritland K (2004) Single-copy, species-transferable microsatellite markers developed from loblolly pine ESTs. Theor Appl Genet 109:361–369. doi:10.1007/s00122-004-1635-7
Mariette S, Cottrell J, Csaikl UM, Goikoechea P, Konig A, Lowe AJ, Van Dam BC, Barreneche T, Bodénès C, Streiff R, Burg K, Groppe K, Munro RC, Tabbener H, Kremer A (2002) Comparison of levels of genetic diversity detected with AFLP and microsatellite markers within and among mixed Q. petraea (matt.) liebl. and Q. robur L. stands. Silvae Genet 51:72–79
Maynard Smith J, Haigh J (1974) The hitch-hiking effect of a favourable gene. Genet Rese 23:23–35. doi:10.1017/S0016672300014634
McShea WJ, Healy WM, Devers P, Fearer T, Koch FH, Stauffer D, Waldon J (2007) Forestry matters: decline of oaks will impact wildlife in hardwood forests. J Wildl Manage 71:1717–1728. doi:10.219
Moran EV, Willis J, Clark JS (2012) Genetic evidence for hybridization in red oaks (Quercus sect. Lobatae, Fagaceae). Am J Bot 99:92–100. doi:10.3732/ajb.11000233/2006-169
Muir G, Schlötterer C (2005) Evidence for shared ancestral polymorphism rather than recurrent gene flow at microsatellite loci differentiating two hybridising oaks (Quercus spp.). Mol Ecol 14:549–561. doi:10.1111/j.1365-294X.2004.02418.x
Oldfield S, Eastwood A (2007) The red list of oaks. Fauna & Flora International Cambridge, UK
Ostfeld RS, Jones CG, Wolff JO (1996) Of mice and mast. Bioscience 46:323–330. doi:10.2307/1312946
Pompanon F, Bonin A, Bellemain E, Taberlet P (2005) Genotyping errors: causes, consequences and solutions. Nat Rev Genet 6:847–859. doi:10.1038/nrg1707
Raymond M, Rousset F (1995) GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225. doi:10.2307/2409177
Rozen S, Skaletsky HJ (2000) PRIMER3 on the www for general users and for biologist programmers. In: Krawertz S, Misener S (eds) Bioinformatic methods and protocols: Methods in molecular biology. Humana Press, Totowa, pp 365–386
Scotti-Saintagne C, Mariette S, Porth I, Goicoechea PG, Barreneche T, Bodénès C, Burg K, Kremer A (2004) Genome scanning of interspecific differentiation between two closely related oak species (Quercus robur L. and Q. petraea (matt.) liebl.). Genetics 168:1615–1626. doi:10.1534/genetics.104.026849
Slate J, Marshall TC, Pemberton JM (2000) A retrospective assessment of the accuracy of the paternity inference program CERVUS. Mol Ecol 9:801–808. doi:10.1046/j.1365-294x.2000.00930.x
Sork VL, Huang S, Wiener E (1993) Macrogeographic and fine-scale genetic structure in a North American oak species, Quercus rubra L. Ann Sci For 50(suppl 1):261s–270s
Sridha S, Wu K (2006) Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J 46:124–133. doi:10.1111/j.1365-313X.2006.02678.x
Steinkellner H, Lexer C, Turetschek E, Glössl J (2003) Conservation of (GA)n microsatellite loci between Quercus species. Mol Ecol 6:1189–1194. doi:10.1046/j.1365-294X.1997.00288.x
Tomlinson PT, Jensen RJ, Hancock JF (2000) Do whole tree silvic characters indicate hybridization in red oak (Quercus section Lobatae)? Am Midl Nat 143:154–168. doi:10.1674/0003-0031(2000)143[0154:dwtsci]2.0.co;2
Tran L-SP, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci U S A 104:20623–20628. doi:10.1073/pnas.0706547105
United Nations Economic Commission for Europe/Food and Agriculture Organization of the United Nations (UNECE/FAO) (2011) Forest products annual market review 2010–2011. Geneva timber and forest study paper 27. United Nations Publications, Geneva
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Microchecker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. doi:10.1111/j.1471-8286.2004.00684.x
van Oosterhout C, Weetman D, Hutchinson WF (2006) Estimation and adjustment of microsatellite null alleles in nonequilibrium populations. Mol Ecol Notes 6:255–256. doi:10.1111/j.1471-8286.2005.01082.x
Vasemägi A, Nilsson J, Primmer CR (2005) Expressed sequence tag-linked microsatellites as a source of gene-associated polymorphisms for detecting signatures of divergent selection in Atlantic salmon (Salmo salar L.). Mol Biol Evol 22:1067–1076. doi:10.1093/molbev/msi09
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. doi:10.2307/2408641
Zane L, Bargelloni L, Patarnello T (2002) Strategies for microsatellite isolation: a review. Mol Ecol 11:1–16. doi:10.1046/j.0962-1083.2001.01418.x
Zhu X, Raman H, Wu H, Lemerle D, Burrows G, Stanton R (2012) Development of SSR markers for menetic analysis of silverleaf nightshade (Solanum elaeagnifolium) and related species. Plant Mol Biol Report: 1–7. doi:10.1007/s11105-012-0473-z
Acknowledgements
We thank James Schmierer for his help in sample collection in the Ford Forestry Center and Dr. Andrew Hipp for providing Q. ellipsoidalis, Q. coccinea and Q. velutina samples. Additionally, we thank John Lampereur and Linda Parker for kindly guiding us to Q. rubra stands in the Nicolet-Chequamegon National Forest. We are also grateful to Erin Hickey who assisted in sample collection and marker analyses. Two anonymous referees provided helpful comments on an earlier draft. Funding for the study came from Michigan Technological University start-up funds to Oliver Gailing, the Michigan Technological University Research Excellence fund, the USDA McIntire Stennis fund, the Huron Mountain Wildlife Foundation, the Hanes Trust and the NSF Plant Genome Research program (NSF 1025974). We are also grateful to the Northern Institute of Applied Climate Science for their support.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(XLSX 18 kb)
Rights and permissions
About this article
Cite this article
Sullivan, A.R., Lind, J.F., McCleary, T.S. et al. Development and Characterization of Genomic and Gene-Based Microsatellite Markers in North American Red Oak Species. Plant Mol Biol Rep 31, 231–239 (2013). https://doi.org/10.1007/s11105-012-0495-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-012-0495-6