Abstract
In modern drug discovery process, ADME/Tox properties should be determined as early as possible in the test cascade to allow a timely assessment of their property profiles. To help medicinal chemists in designing new compounds with improved pharmacokinetics, the knowledge of the soft spot position or the site of metabolism (SOM) is needed. In silico methods based on docking, molecular dynamics and quantum chemical calculations can bring us closer to understand drug metabolism and predict drug–drug interactions. We report herein on a combined methodology to explore the site of metabolism prediction of a new cardioactive drug prototype, LASSBio-294 (1), using MetaPrint2D to predict the most likely metabolites, combined with structure-based tools using docking, molecular dynamics and quantum mechanical calculations to predict the binding of the substrate to CYP2C9 enzyme, to estimate the binding free energy and to study the energy profiles for the oxidation of (1). Additionally, the computational study was correlated with a metabolic fingerprint profiling using LC-MS analysis. The results obtained using the computational methods gave valuable information about the probable metabolites of (1) (qualitatively) and also about the important interactions of this lead compound with the amino acid residues of the active site of CYP2C9. Moreover, using a combination of different levels of theory sheds light on the understanding of (1) metabolism by CYP2C9 and its mechanisms. The metabolic fingerprint profiling of (1) has shown that the metabolites founded in highest concentration in different species were metabolites M1, M2 and M3, whereas M8 was found to be a minor metabolite. Therefore, our computational study allowed a qualitative prediction for the metabolism of (1). The approach presented here has afforded new opportunities to improve metabolite identification strategies, mediated by not only CYP2C9 but also other CYP450 family enzymes.

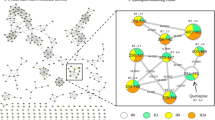
Workflow for metabolic investigation proposed in our work. (a) Proposed computational methods to improve drug metabolism studies using different levels of theory, showing the energy changes from substrate binding to product formation in CYP450-catalyzed drug metabolism. (b) Metabolic fingerprint workflow for complex matrix analysis from different species. Stage I: the preparation of the samples to be analyzed by LC-MS; Stage II: analyze the samples into LC-MS and treat the data; and Stage III: identify and quantify all detectable metabolites produced in vitro by filamentous fungi and discover the similarity across the species using PCA analysis









Similar content being viewed by others
References
Jones BC, Middleton DS, Youdim K (2009) Cytochrome P450 metabolism and inhibition: analysis for drug discovery. Prog Med Chem 47:239–263. doi:10.1016/S0079-6468(08)00206-3
Vistoli G, Pedretti A, Testa B (2008) Assessing drug-likeness–what are we missing? Drug Discov Today 13:285–294. doi:10.1016/j.drudis.2007.11.007
Lewis DFV, Ito Y (2008) Human cytochromes P450 in the metabolism of drugs: new molecular models of enzyme-substrate interactions. Expert Opin Drug Metab Toxicol 4:1181–1186. doi:10.1517/17425250802352412
de Montellano PRO (2010) Cytochrome P450: structure, mechanism, and biochemistry, 3rd edn. Springer, New York
Baranczewski P, Stanczak A, Sundberg K, Svensson R, Wallin A, Jansson J, Garberg P, Postlind H (2006) Introduction to in vitro estimation of metabolic stability and drug interactions of new chemical entities in drug discovery and development. Pharmacol Rep 58:453–472
de Graaf C, Pospisil P, Pos W, Folkers G, Vermeulen NPE (2005) Binding mode prediction of cytochrome p450 and thymidine kinase protein-ligand complexes by consideration of water and rescoring in automated docking. J Med Chem 48:2308–2318. doi:10.1021/jm049650u
Stjernschantz E, Vermeulen NPE, Oostenbrink C (2008) Computational prediction of drug binding and rationalisation of selectivity towards cytochromes P450. Expert Opin Drug Metab Toxicol 4:513–527. doi:10.1517/17425255.4.5.513
Miners JO, Birkett DJ (1998) Cytochrome P4502C9: an enzyme of major importance in human drug metabolism. Br J Clin Pharmacol 45:525–538. doi:10.1046/j.1365-2125.1998.00721.x
Sykes MJ, McKinnon RA, Miners JO (2008) Prediction of metabolism by cytochrome P450 2C9: alignment and docking studies of a validated database of substrates. J Med Chem 51:780–791. doi:10.1021/jm7009793
Wester MR, Yano JK, Schoch GA, Yang C, Griffin KJ, Stout CD, Johnson EF (2004) The structure of human cytochrome P450 2C9 complexed with flurbiprofen at 2.0-A resolution. J Biol Chem 279:35630–35637. doi:10.1074/jbc.M405427200
Usmani KA, Karoly ED, Hodgson E, Rose RL (2004) In vitro sulfoxidation of thioether compounds by human cytochrome P450 and flavin-containing monooxygenase isoforms with particular reference to the CYP2C subfamily. Drug Metab Dispos 32:333–339. doi:10.1124/dmd.32.3.333
Khan MTH (2010) Predictions of the ADMET properties of candidate drug molecules utilizing different QSAR/QSPR modelling approaches. Curr Drug Metab 11(4):285–295. doi:10.2174/138920010791514306
Sun H, Scott DO (2010) Structure-based drug metabolism predictions for drug design. Chem Biol Drug Des 75:3–17. doi:10.1111/j.1747-0285.2009.00899.x
Figueiredo JM, Camara CD, Amarante EG, Miranda ALP, Santos FM, Rodrigues CR, Fraga CAM, Barreiro EJ (2000) Design and synthesis of novel potent antinociceptive agents: methyl-imidazolyl N-acylhydrazone derivatives. Bioorg Med Chem 8:2243–2248. doi:10.1016/S0968-0896(00)00152-8
Zapata-Sudo G, Sudo RT, Maronas PA, Silva GLM, Moreira OR, Aguiar MIS, Barreiro EJ (2003) Thienylhydrazone derivative increases sarcoplasmic reticulum Ca2+ release in mammalian skeletal muscle. Eur J Pharmacol 470:79–85. doi:10.1016/S0014-2999(03)01757-6
Costa DG, da Silva JS, Kummerle AE, Sudo RT, Landgraf SS, Caruso-Neves C, Fraga CAM, de Lacerda Barreiro EJ, Zapata-Sudo G (2010) LASSBio-294, A compound with inotropic and lusitropic activity, decreases cardiac remodeling and improves Ca2(+) influx into sarcoplasmic reticulum after myocardial infarction. Am J Hypertens 23:1220–1227. doi:10.1038/ajh.2010.157
Carlsson L, Spjuth O, Adams S, Glen RC, Boyer S (2010) Use of historic metabolic biotransformation data as a means of anticipating metabolic sites using MetaPrint2D and Bioclipse. BMC Bioinforma 11:362–362. doi:10.1186/1471-2105-11-362
Dewar MJS, Zoebisch EG, Healy EF, Stewart JJP (1985) The development and use of quantum-mechanical molecular-models.76. Am1 - a new general-purpose quantum-mechanical molecular-model. J Am Chem Soc 107:3902–3909. doi:10.1021/ja00299a024
Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1996) A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc 118:2309–2309. doi:10.1021/ja955032e
Oda A, Yamaotsu N, Hirono S (2005) New AMBER force field parameters of heme iron for cytochrome P450s determined by quantum chemical calculations of simplified models. J Comput Chem 26:818–826. doi:10.1002/jcc.20221
Clark AM, Labute P (2007) 2D depiction of protein-ligand complexes. J Chem Inf Model 47:1933–1944. doi:10.1021/ci7001473
Labute P (2008) The generalized Born/volume integral implicit solvent model: estimation of the free energy of hydration using London dispersion instead of atomic surface area. J Comput Chem 29:1693–1698. doi:10.1002/jcc.20933
young dc (2009) computational drug design: a guide for computational and medicinal chemists 1 Har/Cdr edn. Wiley-Interscience, Hoboken. N.J
Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan Y, Wriggers W (2010) Atomic-level characterization of the structural dynamics of proteins. Science 330:341–346. doi:10.1126/science.1187409
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236. doi:10.1021/ja9621760
Paterlini MG, Ferguson DM (1998) Constant temperature simulations using the Langevin equation with velocity Verlet integration. Chem Phys 236:243–252. doi:10.1016/S0301-0104(98)00214-6
SZYBKI (2010). 1.5.1 edn. OpenEye Scientific Software Inc, Santa Fe, NM
Wlodek S, Skillman AG, Nicholls A (2010) Ligand entropy in Gas-Phase. Upon solvation and protein complexation. Fast estimation with Quasi-Newton Hessian. J Chem Theory Comput 6:2140–2152. doi:10.1021/ct100095p
Jaguar (2009) 7.6 edn. Schrodinger, LLC, New York
Braga RC, Tôrres ACB, Persiano CB, Alves RO, Fraga CAM, Barreiro EJ, de Oliveira V (2011) Determination of the cardioactive prototype LASSBio-294 and its metabolites in dog plasma by LC-MS/MS: Application for a pharmacokinetic study. J Pharm Biomed Anal 55:1024–1030. doi:10.1016/j.jpba.2011.02.031
Carneiro EO, Andrade CH, Braga RC, Torres ACB, Alves RO, Liao LM, Fraga CAM, Barreiro EJ, de Oliveira V (2010) Structure-based prediction and biosynthesis of the major mammalian metabolite of the cardioactive prototype LASSBio-294. Bioorg Med Chem Lett 20:3734–3736. doi:10.1016/j.bmcl.2010.04.073
Ts P, Castillo S, Villar-Briones A, Oresic M (2010) MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinforma 11:395. doi:10.1186/1471-2105-11-395
Castro-Perez JM (2007) Current and future trends in the application of HPLC-MS to metabolite-identification studies. Drug Discov Today 12:249–256. doi:10.1016/j.drudis.2007.01.007
Bathelt CM, Zurek J, Mulholland AJ, Harvey JN (2005) Electronic structure of compound I in human isoforms of cytochrome P450 from QM/MM modeling. J Am Chem Soc 127:12900–12908. doi:10.1021/ja0520924
Czodrowski P, Kriegl JM, Scheuerer S, Fox T (2009) Computational approaches to predict drug metabolism. Expert Opin Drug Metab Toxicol 5:15–27. doi:10.1517/17425250802568009
Boyer S, Arnby CH, Carlsson L, Smith J, Stein V, Glen RC (2007) Reaction site mapping of xenobiotic biotransformations. J Chem Inf Model 47:583–590
Rydberg P, Vasanthanathan P, Oostenbrink C, Olsen L (2009) Fast prediction of cytochrome P450 mediated drug metabolism. ChemMedChem 4:2070–2079. doi:10.1002/cmdc.200900363
Park J-Y, Harris D (2003) Construction and assessment of models of CYP2E1: predictions of metabolism from docking, molecular dynamics, and density functional theoretical calculations. J Med Chem 46:1645–1660. doi:10.1021/jm020538a
Lewis DFV (2000) On the recognition of mammalian microsomal cytochrome P450 substrates and their characteristics - Towards the prediction of human P450 substrate specificity and metabolism. Biochem Pharmacol 60:293–306. doi:10.1016/S0006-2952(00)00335-X
Fraga AGM, Fraga CAM, Barreiro EJ, Romeiro NC Perfil metabolico in silico de Prototipo N-Acilidrazonico Cardioativo. In: 30th Reunião Anual da Sociedade Brasileira de Química, Águas de Lindóia - SP, 2007. Abstracts of Papers. Sociedade Brasileira de Química, pp MD-053
Tarcsay A, Rb K, GrM K (2010) Site of metabolism prediction on cytochrome P450 2C9: a knowledge-based docking approach. J Comput-Aided Mol Des 24:399–408. doi:10.1007/s10822-010-9347-3
de Groot MJ, Alex AA, Jones BC (2002) Development of a combined protein and pharmacophore model for cytochrome P450 2C9. J Med Chem 45:1983–1993. doi:jm0110791[pii]
Clodfelter KH, Waxman DJ, Vajda S (2006) Computational solvent mapping reveals the importance of local conformational changes for broad substrate specificity in mammalian cytochromes P450. Biochemistry 45:9393–9407. doi:10.1021/bi060343v
Polgar T, Menyhard DK, Keseru GM (2007) Effective virtual screening protocol for CYP2C9 ligands using a screening site constructed from flurbiprofen and S-warfarin pockets. J Comput-Aided Mol Des 21:539–548. doi:10.1007/S10822-007-9137-8
Williams PA, Cosme J, Ward A, Angove HC, Matak Vinkoviƒá D, Jhoti H (2003) Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature 424:464–468. doi:10.1038/nature01862
Rydberg P, Rod TH, Olsen L, Ryde U (2007) Dynamics of water molecules in the active-site cavity of human cytochromes P450. J Phys Chem B 111:5445–5457. doi:10.1021/jp070390c
Santos R, Hritz J, Oostenbrink C (2010) Role of water in molecular docking simulations of cytochrome P450 2D6. J Chem Inf Model 50:146–154. doi:10.1021/ci900293e
Mulholland AJ (2005) Modelling enzyme reaction mechanisms, specificity and catalysis. Drug Discov Today 10:1393–1402. doi:10.1016/S1359-6446(05)03611-1
Shaik S, Cohen S, Wang Y, Chen H, Kumar D, Thiel W (2010) P450 enzymes: their structure, reactivity, and selectivity-modeled by QM/MM calculations. Chem Rev 110:949–1017. doi:10.1021/cr900121s
Shaik S, Milko P, Schyman P, Usharani D, Chen H (2011) Trends in aromatic oxidation reactions catalyzed by Cytochrome P450 Enzymes: a valence bond modeling. J Chem Theory Comput 7:327–339. doi:10.1021/ct100554g
Schoneboom JC, Lin H, Reuter N, Thiel W, Cohen S, Ogliaro F, Shaik S (2002) The elusive oxidant species of cytochrome P450 enzymes: characterization by combined quantum mechanical/molecular mechanical (QM/MM) calculations. J Am Chem Soc 124:8142–8151. doi:ja026279w[pii]
Kellner DG, Hung SC, Weiss KE, Sligar SG (2002) Kinetic characterization of compound I formation in the thermostable cytochrome P450 CYP119. J Biol Chem 277:9641–9644. doi:10.1074/jbc.C100745200
Crestoni ME, Fornarini S, Lanucara F (2009) Oxygen-atom transfer by a naked manganese(V)-Oxo-Porphyrin complex reveals axial ligand effect. Chem-Eur J 15:7863–7866. doi:10.1002/Chem.200901361
Chiavarino B, Cipollini R, Crestoni ME, Fornarini S, Lanucara F, Lapi A (2008) Probing the Compound I-like reactivity of a bare high-valent oxo iron porphyrin complex: the oxidation of tertiary amines. J Am Chem Soc 130:3208–3217. doi:10.1021/ja077286t
Lonsdale R, Ranaghan KE, Mulholland AJ (2010) Computational enzymology. Chem Commun 46:2354–2372. doi:10.1039/B925647d
Rydberg P, Ryde U, Olsen L (2008) Sulfoxide, sulfur, and nitrogen oxidation and dealkylation by Cytochrome P450. J Chem Theor Comput 4:1369–1377. doi:10.1021/ct800101v
Watanabe Y (2001) Alternatives to the oxoferryl porphyrin cation radical as the proposed reactive intermediate of cytochrome P450: two-electron oxidized Fe(III) porphyrin derivatives. J Biol Inorg Chem 6:846–856. doi:10.1007/s007750100278
Porro CS, Sutcliffe MJ, de Visser SP (2009) Quantum mechanics/molecular mechanics studies on the sulfoxidation of dimethyl sulfide by compound I and compound 0 of cytochrome P450: which is the better oxidant? J Phys Chem A 113:11635–11642. doi:10.1021/jp9023926
Bathelt CM, Mulholland AJ, Harvey JN (2008) QM/MM modeling of benzene hydroxylation in human cytochrome P450 2C9. J Phys Chem A 112:13149–13156. doi:10.1021/jp8016908
de Visser SP, Shaik S (2003) A proton-shuttle mechanism mediated by the porphyrin in benzene hydroxylation by cytochrome P450 enzymes. J Am Chem Soc 125:7413–7424. doi:10.1021/Ja034142f
Costa EMDB, Pimenta FC, Luz WC, de Oliveira V (2008) Selection of filamentous fungi of the Beauveria genus able to metabolize quercetin like mammalian cells. Braz J Microbiol 39:405–408. doi:10.1590/S1517-83822008000200036
Pazini F, Menegatti R, Sabino JR, Andrade CH, Neves G, Rates SMK, Noel F, Fraga CAM, Barreiro EJ, de Oliveira V (2010) Design of new dopamine D2 receptor ligands: biosynthesis and pharmacological evaluation of the hydroxylated metabolite of LASSBio-581. Bioorg Med Chem Lett 20:2888–2891. doi:10.1016/J.Bmcl.2010.03.034
Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G (2005) METLIN: a metabolite mass spectral database. Ther Drug Monit 27:747–751
Acknowledgments
The authors would like to thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Fundação de Amparo à Pesquisa do Estado de Goiás (FAPEG) for financial support and fellowships.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Braga, R.C., Alves, V.M., Fraga, C.A.M. et al. Combination of docking, molecular dynamics and quantum mechanical calculations for metabolism prediction of 3,4-methylenedioxybenzoyl-2-thienylhydrazone. J Mol Model 18, 2065–2078 (2012). https://doi.org/10.1007/s00894-011-1219-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-011-1219-9