Abstract
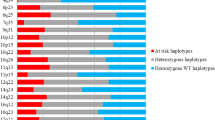
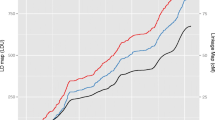
One of the many potential uses of the HapMap project is its application to the investigation of complex disease aetiology among a wide range of populations. This study aims to assess the transferability of HapMap SNP data to the Spanish population in the context of cancer research. We have carried out a genotyping study in Spanish subjects involving 175 candidate cancer genes using an indirect gene-based approach and compared results with those for HapMap CEU subjects. Allele frequencies were very consistent between the two samples, with a high positive correlation (R) of 0.91 (P<<1×10−6). Linkage disequilibrium patterns and block structures across each gene were also very similar, with disequilibrium coefficient (r 2) highly correlated (R=0.95, P<<1×10−6). We found that of the 21 genes that contained at least one block larger than 60 kb, nine (ATM, ATR, BRCA1, ERCC6, FANCC, RAD17, RAD50, RAD54B and XRCC4) belonged to the GO category “DNA repair”. Haplotype frequencies per gene were also highly correlated (mean R=0.93), as was haplotype diversity (R=0.91, P<<1×10−6). “Yin yang” haplotypes were observed for 43% of the genes analysed and 18% of those were identical to the ancestral haplotype (identified in Chimpazee). Finally, the portability of tagSNPs identified in the HapMap CEU data using pairwise r 2 thresholds of 0.8 and 0.5 was assessed by applying these to the Spanish and current HapMap data for 66 genes. In general, the HapMap tagSNPs performed very well. Our results show generally high concordance with HapMap data in allele frequencies and haplotype distributions and confirm the applicability of HapMap SNP data to the study of complex diseases among the Spanish population.






Similar content being viewed by others
References
Abecasis GR, Noguchi E, Heinzmann A, Traherne JA, Bhattacharyya S, Leaves NI, Anderson GG, Zhang Y, Lench NJ, Carey A, Cardon LR, Moffatt MF, Cookson WO (2001) Extent and distribution of linkage disequilibrium in three genomic regions. Am J Hum Genet 68:191–197
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265
Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I (2001) Controlling the false discovery rate in behavior genetics research. Behavl Brain Res 125:279–284
Bonnen PE, Wang PJ, Kimmel M, Chakraborty R, Nelson DL (2002) Haplotype and linkage disequilibrium architecture for human cancer-associated genes. Genome Res 12:1846–1853
Cannon GB (1963) The effects of natural selection on linkage disequilibrium and relative fitness in experimental populations of Drosophila melanogaster. Genetics 48:1201–1216
Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA (2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74:106–120
Clark AG (2003) Finding genes underlying risk of complex disease by linkage disequilibrium mapping. Curr Opin Genetics Dev 13:296–302
Clark AG, Weiss KM, Nickerson DA, Taylor SL, Buchanan A, Stengard J, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, Sing CF (1998) Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am J Hum Genet 63:595–612
Costas J, Salas A, Phillips C, Carracedo A (2005) Human genome-wide screen of haplotype-like blocks of resuced diversity. Gene 349:219–225
Crawford DC, Carlson CS, Rieder MJ, Carrington DP, Yi Q, Smith JD, Eberle MA, Kruglyak L, Nickerson DA (2004) Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am J Hum Genet 74:610–622
Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES (2001) High-resolution haplotype structure in the human genome. Nat Genet 29:229–232
Dawson E, Abecasis GR, Bumpstead S, Chen Y, Hunt S, Beare DM, Pabial J, Dibling T, Tinsley E, Kirby S, Carter D, Papaspyridonos M, Livingstone S, Ganske R, Lohmussaar E, Zernant J, Tonisson N, Remm M, Magi R, Puurand T, Vilo J, Kurg A, Rice K, Deloukas P, Mott R, Metspalu A, Bentley DR, Cardon LR, Dunham I (2002) A first-generation linkage disequilibrium map of human chromosome 22. Nature 418:544–548
De la Vega FM, Lazaruk KD, Rhodes MD, Wenz MH (2005) Assessment of two flexible and compatible SNP genotyping platforms: TaqMan SNP Genotyping Assays and the SNPlex Genotyping System. Mutat Res 573:111–135
Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4:3
Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229
Goldstein DB (2001) Islands of linkage disequilibrium. Nat Genet 29:109–111
Gonzalez-Neira A, Calafell F, Navarro A, Lao O, Cann H, Comas D, Bertranpetit J (2004) Geographic stratification of linkage disequilibrium: a worldwide population study in a region of chromosome 22. Hum Genomics 1:399–409
Hartl DA, Clark AG (1997) Principle of population genetics. Sunderland, MA
Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR (2005) Whole-genome patterns of common DNA variation in three human populations. Science 307:1072–1079
Hosack DA, Dennis G Jr, Sherman BT, Lane HC, Lempicki RA (2003) Identifying biological themes within lists of genes with EASE. Genome Biol 4:70
Hudson R (1990) Gene genealogies and the coalescent process. Oxf Surv Evol Biol 7:1
Hurst LD, Pal C (2001) Evidence for purifying selection acting on silent sites in BRCA1. Trends Genet 17:62–65
Huttley GA, Smith MW, Carrington M, O’Brien SJ (1999) A scan for linkage disequilibrium across the human genome. Genetics 152:1711–1722
Janosikova B, Zavadakova P, Kozich V (2005) Single-nucleotide polymorphisms in genes relating to homocysteine metabolism: how applicable are public SNP databases to a typical European population?. Eur J Hum Genet 13:86–95
Jorde LB (2000) Linkage disequilibrium and the search for complex disease genes. Genome Res 10:1435–1444
Kamatani N, Sekine A, Kitamoto T, Iida A, Saito S, Kogame A, Inoue E, Kawamoto M, Harigai M, Nakamura Y (2004) Large-scale single-nucleotide polymorphism (SNP) and haplotype analyses, using dense SNP Maps, of 199 drug-related genes in 752 subjects: the analysis of the association between uncommon SNPs within haplotype blocks and the haplotypes constructed with haplotype-tagging SNPs. Am J Hum Genet 75:190–203
Ke X, Hunt S, Tapper W, Lawrence R, Stavrides G, Ghori J, Whittaker P, Collins A, Morris AP, Bentley D, Cardon LR, Deloukas P (2004) The impact of SNP density on fine-scale patterns of linkage disequilibrium. Hum Mol Genet 13:577–588
Kelly JK, Wade MJ (2000) Molecular evolution near a two-locus balanced polymorphism. J Theor Biol 204:83–101
Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247
Liu Y, Yoshimura K, Hanaoka T, Ohnami S, Kohno T, Yoshida T, Sakamoto H, Sobue T, Tsugane S (2005) Association of habitual smoking and drinking with single nucleotide polymorphism (SNP) in 40 candidate genes: data from random population-based Japanese samples. J Hum Genet 50:62–68
Long JR, Zhao LJ, Liu PY, Lu Y, Dvornyk V, Shen H, Liu YJ, Zhang YY, Xiong DH, Xiao P, Deng HW (2004) Patterns of linkage disequilibrium and haplotype distribution in disease candidate genes. BMC Genetics 5:11
McCarthy JJ, Parker A, Salem R, Moliterno DJ, Wang Q, Plow EF, Rao S, Shen G, Rogers WJ, Newby LK, Cannata R, Glatt K, Topol EJ, GeneQuest I (2004) Large scale association analysis for identification of genes underlying premature coronary heart disease: cumulative perspective from analysis of 111 candidate genes. J Med Genet 41:334–341
Mueller JC, Lohmussaar E, Magi R, Remm M, Bettecken T, Lichtner P, Biskup S, Illig T, Pfeufer A, Luedemann J, Schreiber S, Pramstaller P, Pichler I, Romeo G, Gaddi A, Testa A, Wichmann HE, Metspalu A, Meitinger T (2005) Linkage disequilibrium patterns and tagSNP transferability among European populations. Am J Hum Genet 76:387–398
Nei M, Tajima F (1981) DNA polymorphism detectable by restriction endonucleases. Genetics 97:145–163
Nejentsev S, Godfrey L, Snook H, Rance H, Nutland S, Walker NM, Lam AC, Guja C, Ionescu-Tirgoviste C, Undlien DE, Ronningen KS, Tuomilehto-Wolf E, Tuomilehto J, Newport MJ, Clayton DG, Todd JA (2004) Comparative high-resolution analysis of linkage disequilibrium and tag single nucleotide polymorphisms between populations in the vitamin D receptor gene. Hum Mol Genet 13:1633–1639
Nyholt DR (2004) A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 74:765–769
Oliphant A, Barker DL, Stuelpnagel JR, Chee MS (2002) BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques Suppl: 56–58
Pritchard JK, Cox NJ (2002) The allelic architecture of human disease genes: common disease–common varient or not? Hum Mol Genet 11:2417–2423
Sawyer SL, Mukherjee N, Pakstis AJ, Feuk L, Kidd JR, Brookes AJ, Kidd KK (2005) Linkage disequilibrium patterns vary substantially among populations. Eur J Hum Genet 13:677–686
Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, Stanley SE, Jiang R, Messer CJ, Chew A, Han JH, Duan J, Carr JL, Lee MS, Koshy B, Kumar AM, Zhang G, Newell WR, Windemuth A, Xu C, Kalbfleisch TS, Shaner SL, Arnold K, Schulz V, Drysdale CM, Nandabalan K, Judson RS, Ruano G, Vovis GF (2001) Haplotype variation and linkage disequilibrium in 313 human genes. Science 293:489–493
The Gene Ontology C (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25:25–29
The International HapMap Consortium (2003) The International HapMap Project. Nature 426:789–796
The International HapMap Consortium (2004) Integrating ethics and science in the International HapMap Project. Nat Rev Genet 5:467–475
Thompson D, Stram D, Goldgar D, Witte JS (2003) Haplotype tagging single nucleotide polymorphisms and association studies. Hum Hered 56:48–55
Wall JD, Pritchard JK (2003) Assessing the performance of the haplotype block model of linkage disequilibrium. Am J Hum Genet 73:502–515
Zavattari P, Deidda E, Whalen M, Lampis R, Mulargia A, Loddo M, Eaves I, Mastio G, Todd JA, Cucca F (2000) Major factors influencing linkage disequilibrium by analysis of different chromosome regions in distinct populations: demography, chromosome recombination frequency and selection. Hum Mol Genet 9:2947–2957
Zhang J, Rowe WL, Clark AG, Buetow KH (2003) Genomewide distribution of high-frequency, completely mismatching SNP haplotype pairs observed to be common across human populations. Am J Hum Genet 73:1073–1081
Web site references
http://www.hapmap.org/ HapMap
http://www.ensemble.org/ Ensembl homepage
http://www.geneontology.org/GO website
Acknowledgements
EB and LPF are funded by the Comunidad Autónoma de Madrid and by the Spanish Ministry of Science and Technology (MCT), respectively. We thank Christian Torrenteras for Illumina platform support and Fatima Mercadillo and for her expert technical skills. We would also like to thank Christopher Philips and Beatriz Sobrino for their technical support with the SNPlex genotyping platform as well as Jorge Amigo for his assistance with the Genotyping Data Formatter software used to parse SNPlex data and to control genotyping errors. The National Genotyping Centre (CeGen) is funded by the Genome Spain Foundation. This study was partially supported by BFI2003-03852, PI020919, PI041313 and PGIDIT02PXIC20804PN.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Ribas, G., González-Neira, A., Salas, A. et al. Evaluating HapMap SNP data transferability in a large-scale genotyping project involving 175 cancer-associated genes. Hum Genet 118, 669–679 (2006). https://doi.org/10.1007/s00439-005-0094-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-005-0094-9