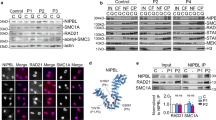
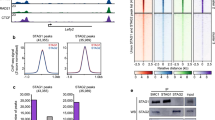
Abstract
Drosophila Nipped-B is an essential protein that has multiple functions. It facilitates expression of homeobox genes and is also required for sister chromatid cohesion. Nipped-B is conserved from yeast to man, and its orthologs also play roles in deoxyribonucleic acid repair and meiosis. Mutation of the human ortholog, Nipped-B-Like (NIPBL), causes Cornelia de Lange syndrome (CdLS), associated with multiple developmental defects. The Nipped-B protein family is required for the cohesin complex that mediates sister chromatid cohesion to bind to chromosomes. A key question, therefore, is whether the Nipped-B family regulates gene expression, meiosis, and development by controlling cohesin. To gain insights into Nipped-B’s functions, we compared the effects of several Nipped-B mutations on gene expression, sister chromatid cohesion, and meiosis. We also examined association of Nipped-B and cohesin with somatic and meiotic chromosomes by immunostaining. Missense Nipped-B alleles affecting the same HEAT repeat motifs as CdLS-causing NIPBL mutations have intermediate effects on both gene expression and mitotic chromatid cohesion, linking these two functions and the role of NIPBL in human development. Nipped-B colocalizes extensively with cohesin on chromosomes in both somatic and meiotic cells and is present in soluble complexes with cohesin subunits in nuclear extracts. In meiosis, Nipped-B also colocalizes with the synaptonemal complex and contributes to maintenance of meiotic chromosome cores. These results support the idea that direct regulation of cohesin function underlies the diverse functions of Nipped-B and its orthologs.









Similar content being viewed by others
References
Anderson LK, Royer SM, Page SL, McKim KS, Lai A, Lilly MA, Hawley RS (2005) Juxtaposition of C(2)M and the transverse filament protein C(3)G within the central region of Drosophila synaptonemal complex. Proc Natl Acad Sci USA 102:4482–4487
Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K (2003) ATP hydrolysis is required for cohesin’s association with chromosomes. Curr Biol 13:1941–1953
Bannister LA, Reinholdt LG, Munroe RJ, Schimenti JC (2004) Positional cloning and characterization of mouse mei8, a disrupted allele of the meiotic cohesin Rec8. Genesis 40:184–194
Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC (2001) Requirement of heterochromatin for cohesion at centromeres. Science 294:2539–2542
Bernard P, Drogat J, Maure JF, Dheur S, Vaur S, Genier S, Javerzat JP (2006) A screen for cohesion mutants uncovers Ssl3, the fission yeast counterpart of the cohesin loading factor Scc4. Curr Biol 16:875–881
Bickel SE, Wyman DW, Orr-Weaver TL (1997) Mutational analysis of the Drosophila sister-chromatid cohesion protein ORD and its role in the maintenance of centromeric cohesion. Genetics 146:1319–1331
Chan RC, Chan A, Jeon M, Wu TF, Pasqualone D, Rougvie AE, Meyer BJ (2003) Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature 423:1002–1009
Chang CR, Wu CS, Hom Y, Gartenberg MR (2005) Targeting of cohesin by transcriptionally silent chromatin. Genes Dev 19:3031–3042
Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K (2000) Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5:243–254
Cummings WJ, Merino ST, Young KG, Li L, Johnson CW, Sierra EA, Zolan ME (2002) The Coprinus cinereus adherin Rad9 functions in Mre11-dependent DNA repair, meiotic sister-chromatid cohesion, and meiotic homolog pairing. Proc Natl Acad Sci USA 99:14958–14963
Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodriguez C, Arnedo M, Loeys B, Kline AD, Wilson M, Lillquist K, Siu V, Ramos FJ, Musio A, Jackson LS, Dorsett D, Krantz ID (2007) Mutations in cohesin complex members Smc3 and Smc1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet 80:485–494
Dobie KW, Kennedy CD, Velasco VM, McGrath TL, Weko J, Patterson RW, Karpen GH (2001) Identification of chromosome inheritance modifiers in Drosophila melanogaster. Genetics 157:1623–1637
Dorsett D (2004) Adherin: key to the cohesin ring and cornelia de Lange syndrome. Curr Biol 14:R834–R836
Dorsett D (2007) Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma 116:1–13
Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K (2005) Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development 132:4743–4753
Eijpe M, Heyting C, Gross B, Jessberger R (2000) Association of mammalian SMC1 and SMC3 proteins with meiotic chromosomes and synaptonemal complexes. J Cell Sci 113:673–682
Fanti L, Giovinazzo G, Berloco M, Pimpinelli S (1998) The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell 2:527–538
Fisher RA (1922) On the interpretation of χ2 from contingency tables, and the calculation of P. J R Stat Soc 85:87–94
Furuya K, Takahashi K, Yanagida M (1998) Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev 12:3408–3418
Gause M, Morcillo P, Dorsett D (2001) Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol Cell Biol 21:4807–4817
Gandhi R, Gillespie PJ, Hirano T (2006) Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol 16:2406–2417
Gillespie PJ, Hirano T (2004) Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol 14:1598–1603
Gillis LA, McCallum J, Kaur M, DeScipio C, Yaeger D, Mariani A, Kline AD, Li HH, Devoto M, Jackson LG, Krantz ID (2004) NIPBL mutational analysis in 120 individuals with Cornelia de Lange syndrome and evaluation of genotype-phenotype correlations. Am J Hum Genet 75:610–623
Hagstrom KA, Meyer BJ (2003) Condensin and cohesin: more than chromosome compactor and glue. Nat Rev Genet 4:520–534
Hartman T, Stead K, Koshland D, Guacci V (2000) Pds5p is an essential chromosomal protein required for both sister chromatid cohesin and condensation in Saccharomyces cerevisiae. J Cell Biol 151:613–626
Heidmann D, Horn S, Heidmann S, Schleiffer A, Nasmyth K, Lehner CF (2004) The Drosophila meiotic kleisin C(2)M functions before the meiotic divisions. Chromosoma 113:177–187
Hirano T (2006) At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol 7:311–322
Huang CE, Milutinovich M, Koshland D (2005) Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Phil Trans R Soc Lond B Biol Sci 360:537–542
Khetani RS, Bickel SE (2007) Regulation of meiotic cohesion and chromosome core morphogenesis during pachytene in Drosophila oocytes. J Cell Sci 120:3123–3137
Kitajima TS, Kawashima SA, Watanabe Y (2004) The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427:510–517
Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K (1999) A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98:91–103
Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG (2004) Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet 36:631–635
Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM (2006) Wapl controls the dynamic association of cohesin with chromatin. Cell 127:955–967
Lechner MS, Schultz DC, Negorev D, Maul GG, Rauscher FJ 3rd (2005) The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem Biophys Res Commun 331:929–937
Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430:573–578
Losada A (2007) Cohesin regulation: fashionable ways to wear a ring. Chromosoma 116:321–329
Losada A, Yokochi T, Hirano T (2005) Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci 118:2133–2141
Manheim EA, McKim KS (2003) The synaptonemal complex component C(2)M regulates meiotic crossing over in Drosophila. Curr Biol 13:276–285
McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K (2005) Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol 3:e86
Michaelis C, Ciosk R, Nasmyth K (1997) Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35–45
Miyake N, Visser R, Kinoshita A, Yoshiura K, Niikawa N, Kondoh T, Matsumoto N, Harada N, Okamoto N, Sonoda T, Naritomi K, Kaname T, Chinen Y, Tonoki H, Kurosawa K (2005) Four novel NIPBL mutations in Japanese patients with Cornelia de Lange syndrome. Am J Med Genet A 135:103–105
Morcillo P, Rosen C, Dorsett D (1996) Genes regulating the remote wing margin enhancer in the Drosophila cut locus. Genetics 144:1143–1154
Murphy TD, Karpen GH (1995) Interactions between the nod + kinesin-like gene and extracentromeric sequences are required for transmission of a Drosophila minichromosome. Cell 81:139–148
Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L (2006) X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet 38:528–530
Myster SH, Wang F, Cavallo R, Christian W, Bhotika S, Anderson CT, Peifer M (2004) Genetic and bioinformatic analysis of 41C and the 2R heterochromatin of Drosophila melanogaster: a window on the heterochromatin-euchromatin junction. Genetics 166:807–822
Nasmyth K, Haering CH (2005) The structure and function of SMC and kleisin complexes. Annu Rev Biochem 74:595–648
Neuwald AF, Hirano T (2000) HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res 10:1445–1452
Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y (2002) Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol 4:89–93
Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K (2000) Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol 10:1557–1564
Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J (2001) A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev 15:1349–1360
Pasierbek P, Fodermayr M, Jantsch V, Jantsch M, Schweizer D, Loidl J (2003) The Caenorhabditis elegans SCC-3 homologue is required for meiotic synapsis and for proper chromosome disjunction in mitosis and meiosis. Exp Cell Res 289:245–255
Revenkova E, Jessberger R (2006) Shaping meiotic prophase chromosomes: cohesins and synaptonemal complex proteins. Chromosoma 115:235–240
Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R (2004) Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol 6:555–562
Rollins RA, Morcillo P, Dorsett D (1999) Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152:577–593
Rollins RA, Korom M, Aulner N, Martens A, Dorsett D (2004) Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol 24:3100–3111
Salic A, Waters JC, Mitchison TJ (2004) Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell 118:567–578
Schoumans J, Wincent J, Barbaro M, Djureinovic T, Maguire P, Forsberg L, Staaf J, Thuresson AC, Borg A, Nordgren A, Malm G, Anderlid BM (2007) Comprehensive mutational analysis of a cohort of Swedish Cornelia de Lange syndrome patients. Eur J Hum Genet 15:143–149
Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, Rollins RA, Erdjument-Bromage H, Tempst P, Benard CY, Hekimi S, Newbury SF, Strachan T (2006) Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol 4:e242
Seitz LC, Tang K, Cummings WJ, Zolan ME (1996) The rad9 gene of Coprinus cinereus encodes a proline-rich protein required for meiotic chromosome condensation and synapsis. Genetics 142:1105–1117
Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI (2002) A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif 25:8–15
Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC (2004) Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol 6:991–996
Tanaka K, Hao Z, Kai M, Okayama H (2001) Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J 20:5779–5790
Tang TT, Bickel SE, Young LM, Orr-Weaver TL (1998) Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S332 protein. Genes Dev 12:3843–3856
Tang Z, Sun Y, Harley SE, Zou H, Yu H (2004) Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc Natl Acad Sci USA 101:18012–18017
Tomonaga T, Nagao K, Kawasaki Y, Furuya K, Murakami A, Morishita J, Yuasa T, Sutani T, Kearsey SE, Uhlmann F, Nasmyth K, Yanagida M (2000) Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev 14:2757–2770
Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T (2004) NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet 36:636–641
Valdeolmillos A, Rufas JS, Suja JA, Vass S, Heck MM, Martinez-A C, Barbero JL (2004) Drosophila cohesins DSA1 and Drad21 persist and colocalize along the centromeric heterochromatin during mitosis. Biol Cell 96:457–462
Valentine G, Wallace YJ, Turner FR, Zolan ME (1995) Pathway analysis of radiation-sensitive meiotic mutants of Coprinus cinereus. Mol Gen Genet 247:169–179
Vass S, Cotterill S, Valdeolmillos AM, Barbero JL, Lin E, Warren WD, Heck MM (2003) Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr Biol 13:208–218
Verni F, Gandhi R, Goldberg ML, Gatti M (2000) Genetic and molecular analysis of wings apart-like (wapl), a gene controlling heterochromatin organization in Drosophila melanogaster. Genetics 154:1693–1710
Warren WD, Steffensen S, Lin E, Coelho P, Loupart M, Cobbe N, Lee JY, McKay MJ, Orr-Weaver T, Heck MM, Sunkel CE (2000) The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr Biol 10:1463–1466
Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM (2006) Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol 16:863–874
Webber HA, Howard L, Bickel SE (2004) The cohesion protein ORD is required for homologue bias during meiotic recombination. J Cell Biol 164:819–829
Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ (2005) Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell 8:949–961
Acknowledgements
The authors thank Steve Myster and Mark Peifer for the EMS-generated Nipped-B mutants, Scott Hawley for C(3)G antibodies, and Gary Karpen for the J21A fly stocks. We also thank Patrick Morcillo for advice on statistical analysis and Ian Krantz and Matt Deardorff for information on human NIPBL mutations. This work was supported by NIH Grants R01 GM055683 (D.D.), R01 GM059354 (S.E.B.); P01 HD058260 (D.D., Project III Director), and March of Dimes FY05-103 (D.D.).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Uhlmann
Gause, Webber, and Misulovin provided equal contributions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Nipped-B Western blots. All blots were probed with the guinea pig anti-Nipped-B serum used in the immunostaining experiments shown in Figs. 5 and 6. The upper left panel shows an overexposed Western blot of whole cell extracts of cultured Sg4 cells, whole adult ovaries, and whole third instar imaginal disks, showing that a band close to the expected size of 237 kDa is the major reacting protein. The right panels are a Western blot of whole-Sg4-cells extracts treated with no RNAi or RNAi directed against Nipped-B (Rollins et al. 2004), showing that the major band is strongly reduced by RNAi. The blot was reprobed for actin as a loading control. The middle left panel is a Western blot of whole early embryos (0 to 12 h after egg laying) from wild-type mothers and mothers heterozygous for the indicated mutant Nipped-B alleles. All lanes contained 100 embryos, and the truncation alleles all give lower levels than the wild type, while the two missense alleles shown have similar levels. The asterisks show very faint double bands consistent with the predicted sizes for the NC7- and NC59-truncated proteins. The bottom panel is a Western blot showing immunoprecipitation of the Kc nuclear extract with the rabbit anti-Nipped-B serum used for the experiment in Fig. 9. The preimmune serum (pre) did not precipitate Nipped-B and left it in the postprecipitation supernatant. The immune serum (imm) precipitates Nipped-B and removes it entirely from the supernatant. All lanes all are from the same gel but have been reordered from the original for clarity (PDF 131 kb)
Supplementary Fig. 2
Locations of Nipped-B wild-type amino acid polymorphisms and EMS-induced mutations. The top is a map of the exons (blue boxes, coding; orange boxes, noncoding) of the previously reported Nipped-B cDNA clone (Rollins et al. 1999; GenBank accession no. AF114160) mapped onto the Drosophila genome scaffold with the putative initiation (ATG) and termination codons (TGA). The positions of polymorphisms that cause changes in amino acid sequence (a–h) and EMS-induced mutations (NC) are indicated. Distances are indicated in kilobases (1K–41K). The table at the bottom lists the differences in amino acid sequence predicted by the wild-type Nipped-B polymorphisms (a–h). cn bw-a is a stock with a wild-type Nipped-B allele kept in our laboratory, and cn bw-b is the wild-type allele used for the mutagenesis (Myster et al. 2004) (PDF 147 kb)
Supplementary Fig. 3
N-terminal alternative splicing of Nipped-B transcripts The maps (a–d) show the first six exons of the standard Nipped-B cDNA (GenBank accession no. AF114160) with alternative splicing variants detected by direct sequencing of PCR-amplified reverse transcription products of total RNA from second instar larvae. a Two closely spaced alternative splice donors occur in exon 1, before the start of the open reading frame (dashed lines). b Splicing from the exon 2 donor site to alternative acceptor site in exon 4, bypassing exon 3, creates a short open reading frame (ATG–TAA). Initiation of translation at the next ATG would produce a Nipped-B protein lacking the conserved N-terminal residues. c Splicing from the exon 2 donor site to the exon 5 acceptor skips exons 3 and 4 but maintains the reading frame. d Splicing of a small intron within exon 6 preserves the reading frame. The predicted protein sequences produced by splicing patterns a–d are given in Supplementary Fig. 4. We do not know if these alternative splicing events occur independently or in combination, although patterns b and c are incompatible with each other (PDF 62.9 kb)
Supplementary Fig. 4
Predicted N-terminal protein sequences for the Nipped-B alternative splicing patterns. The protein sequences for the splicing patterns (a–d) shown in Supplementary Fig. 3 are aligned, with dashes indicating absence of the residues (PDF 488 kb)
Rights and permissions
About this article
Cite this article
Gause, M., Webber, H.A., Misulovin, Z. et al. Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells. Chromosoma 117, 51–66 (2008). https://doi.org/10.1007/s00412-007-0125-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-007-0125-5