Abstract
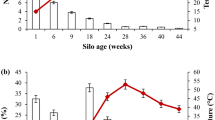
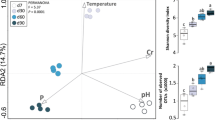
This study investigated the species diversity and substrate utilization patterns of culturable thermophilic bacterial communities in hot aerobic poultry and cattle manure composts by coupling 16S rDNA analysis with Biolog data. Based on the phylogenetic relationships of 16S rDNA sequences, 34 thermophilic (grown at 60°C) bacteria isolated during aerobic composting of poultry manure and cattle manure were classified as Bacillus licheniformis, B. atrophaeus, Geobacillus stearothermophilus, G. thermodenitrificans, Brevibacillus thermoruber, Ureibacillus terrenus, U. thermosphaericus, and Paenibacillus cookii. In this study, B. atrophaeus, Br. thermoruber, and P. cookii were recorded for the first time in hot compost. Physiological profiles of these bacteria, obtained from the Biolog Gram-positive (GP) microplate system, were subjected to principal component analysis (PCA). All isolates were categorized into eight different PCA groups based on their substrate utilization patterns. The bacterial community from poultry manure compost comprised more divergent species (21 isolates, seven species) and utilized more diverse substrates (eight PCA groups) than that from cattle manure compost (13 isolates, five species, and four PCA groups). Many thermophilic bacteria isolated in this study could use a variety of carboxylic acids. Isolate B110 (from poultry manure compost), which is 97.6% similar to U. terrenus in its 16S rDNA sequence, possesses particularly high activity in utilizing a broad spectrum of substrates. This isolate may have potential applications in industry.


Similar content being viewed by others
References
Araujo, A, Ward, OP (1990) Hemicellulases of Bacillus species: preliminary comparative studies on production and properties of mannanases and galactanases. J Appl Bacteriol 68: 253–261
Asai, K, Baik, SH, Kasahara, Y, Moriya, S, Ogasawara, N (2000) Regulation of the transport system for C4-dicarboxylic acids in Bacillus subtilis. Microbiology 146: 263–271
Blanc, M, Marilley, L, Beffa, T, Aragno, M (1999) Rapid identification of heterotrophic, thermophilic, spore-forming bacteria isolated from hot composts. Int J Syst Bacteriol 47: 1246–1248
Blencke, HM, Homuth, G, Ludwig, H, Mader, U, Hecker, M, Stulke, J (2003) Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab Eng 5: 133–149
Carpenter-Boggs, L, Kennedy, AC, Reganold, JP (1998) Use of phospholipid fatty acids and carbon source utilization patterns to track microbial community succession in developing compost. Appl Environ Microbiol 64: 4062–4064
Chauhan, HJ, Domingo, GJ, Jung, HI, Perham, RN (2000) Sites of limited proteolysis in the pyruvate decarboxylase component of the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus and their role in catalysis. Eur J Biochem 267: 7158–7169
Choi, K-H, Dobbs, FC (1999) Comparison of two kinds of Biolog microplates (GN and ECO) in their ability to distinguish among aquatic microbial communities. J Microbiol Methods 36: 203–213
Coleman, ML, Sullivan, MB, Martiny, AC, Steglich, C, Barry, K, DeLong, EF, Chisholm, SW (2006) Genomic islands and the ecology and evolution of Prochlorococcus. Science 311: 1768–1770
Dees, PM, Ghiorse, WC (2001) Microbial diversity in hot synthetic compost as revealed by PCR-amplified rRNA sequences from cultivated isolates and extracted DNA. FEMS Microbiol Ecol 35: 207–216
Ethier, N, Talbot, G., Sygusch, J (1998) Gene cloning, DNA sequencing, and expression of thermostable beta-mannanase from Bacillus stearothermophilus. Appl Environ Microbiol 64: 4428–4432
Fawcett, JK (1954) The semi-micro Kjeldahl method for the determination of nitrogen. J Med Lab Technol 12: 1–22
Felsenstein, J (2004) Phylip (Phylogeny inference package) version 3.62. Distributed by the author. Department of Genetics, University of Washington, Seattle.
Fortnagel, P, Freese, E (1968) Analysis of sporulation mutants. II. Mutants blocked in the citric acid cycle. J Bacteriol 95: 1431–1438
Fritze, D, Pukall, R (2001) Reclassification of bioindicator strains Bacillus subtilis DSM 675 and Bacillus subtilis DSM 2277 as Bacillus atrophaeus. Int J Syst Evol Microbiol 51: 35–37
Gagné, A, Chicoine, M, Morin, A, Houde, A (2001) Phenotypic and genotypic characterization of esterase-producing Ureibacillus thermosphaericus isolated from an aerobic digestor of swine waste. Can J Microbiol 47: 908–915
Halebian, S, Harris, B, Finegold, SM, Rolfe, RD (1981) Rapid method that aids in distinguishing Gram-positive from Gram-negative anaerobic bacteria. J Clin Microbiol 13: 444–448
Herrmann, RF, Shann, JF (1997) Microbial community changes during the composting of municipal solid waste. Microbial Ecol 33: 78–85
Juteau, P, Tremblay, D, Villemur, R, Bisaillon, JG, Beaudet, R (2005) Analysis of the bacterial community inhabiting an aerobic thermophilic sequencing batch reactor (AT-SBR) treating swine waste. Appl Microbiol Biotechnol 66: 115–122
Kelsey, JC (1967) Use of gaseous antimicrobial agents with special reference to ethylene oxide. J Appl Bacteriol 30: 92–100
Kimura, M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120
Kunst, F, Ogasawara, N, Moszer, I, Albertini, AM, Alloni, G, Azevedo, V, Bertero, MG, Bessieres, P, Bolotin, A, Borchert, S, Borriss, R, Boursier, L, Brans, A, Braun, M, Brignell, SC, Bron, S, Brouillet, S, Bruschi, CV, Caldwell, B, Capuano, V, Carter, NM, Choi, SK, Codani, JJ, Connerton, IF, Danchin, A, et al. (1997) The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390: 249–256
Lane, DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (Eds.) Nucleic Acid Techniques in Bacterial Systematics, Wiley, Chichester, pp 115–174
Lebuhn, M, Achouak, W, Schloter, M, Berge, O, Meier, H, Barakat, M, Hartmann, A, Heulin, T (2000) Taxonomic characterization of Ochrobactrum sp. isolates from soil samples and wheat roots, and description of Ochrobactrum tritici sp. nov. and Ochrobactrum grignonense sp. nov. Int J Syst Evol Microbiol 50: 2207–2223
Lin, EC, Iuchi, S (1991) Regulation of gene expression in fermentative and respiratory systems in Escherichia coli and related bacteria. Annu Rev Genet 25: 361–387
Logan, NA, De Clerck, E, Lebbe, L, Verhelst, A, Goris, J, Forsyth, G, Rodriguez-Diaz, M, Heyndrickx, M, De Vos, P (2004) Paenibacillus cineris sp. nov. and Paenibacillus cookii sp. nov., from Antarctic volcanic soils and a gelatin-processing plant. Int J Syst Evol Microbiol 54: 1071–1076
McInroy, SG, Campbell, CD, Haukka, KE, Odee, DW, Sprent, JI, Wang, WJ, Young, JP, Sutherland, JM (1999) Characterization of rhizobia from African acacias and other tropical woody legumes using Biolog and partial 16S rRNA sequencing. FEMS Microbiol Lett 170: 111–117
McMullan, G, Christie, JM, Rahman, TJ, Banat, IM, Ternan, NG, Marchant, R (2004) Habitat, applications and genomics of the aerobic, thermophilic genus Geobacillus. Biochem Soc Trans 32: 214–217
Nelson, DW, Sommers, JE (1996) Total carbon, organic carbon, and organic matter. In: Bigham JM (Ed.) Methods of Soil Analysis: Part 3. Chemical Methods, Soil Science Society of America, Wisconsin, pp 961–1010
Nielsen, JE, Borchert, TV (2000) Protein engineering of bacterial alpha-amylases. Biochim Biophys Acta 1543: 253–274
Niwa, T, Kawamura, Y, Katagiri, Y, Ezaki, T (2005) Lytic enzyme, labiase for a broad range of Gram-positive bacteria and its application to analyze functional DNA/RNA. J Microbiol Methods 61: 251–260
Palys, T, Nakamura, LK, Cohan, FM (1997) Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int J Syst Bacteriol 47: 1145–1156
Sohail, M (1998) A simple and rapid method for preparing genomic DNA from gram-positive bacteria. Mol Biotechnol 10: 191–193
Steger, K, Jarvis, A, Smars, S, Sundh, I (2003) Comparison of signature lipid methods to determine microbial community structure in compost. J Microbiol Mehtods 55: 371–382
Strom, PF (1985) Effect of temperature on bacterial species diversity in thermophilic solid-waste composting. Appl Environ Microbiol 50: 899–905
Tanaka, K, Kobayashi, K, Ogasawara, N (2003) The Bacillus subtilis YufLM two-component system regulates the expression of the malate transporters MaeN (YufR) and YflS, and is essential for utilization of malate in minimal medium. Microbiology 149: 2317–2329
Thambirajah, JJ, Zulkali, MD, Hashim, MA (1995) Microbiological and biochemical changes during the composting of oil palm empty-fruit-bunches. Effect of nitrogen supplementation on the substrate. Biores Technol 52: 133–144
Veith, B, Herzberg, C, Steckel, S, Feesche, J, Maurer, KH, Ehrenreich, P, Baumer, S, Henne, A, Liesegang, H, Merkl, R, Ehrenreich, A, Gottschalk, G (2004) The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J Mol Microbiol Biotechnol 7: 204–211
Zhang, YC, Ronimus, RS, Turner, N, Zhang, Y, Morgan, HW (2002) Enumeration of thermophilic Bacillus species in composts and identification with a Random Amplification Polymorphic DNA (RAPD) protocol. Syst Appl Microbiol 25: 618–626
Author information
Authors and Affiliations
Corresponding author
Additional information
The first two authors contributed equally to this work.
Rights and permissions
About this article
Cite this article
Wang, CM., Shyu, CL., Ho, SP. et al. Species Diversity and Substrate Utilization Patterns of Thermophilic Bacterial Communities in Hot Aerobic Poultry and Cattle Manure Composts. Microb Ecol 54, 1–9 (2007). https://doi.org/10.1007/s00248-006-9139-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-006-9139-4