Abstract
Background
Recently, bacterial endosymbiont, including Wolbachia and Microsporidia were found to limit the infection of Anopheles mosquitoes with Plasmodium falciparum. This study aimed to investigate the natural presence of key transmission-blocking endosymbionts in Anopheles gambiae and Anopheles coluzzii in Southern Benin.
Methods
The present study was conducted in seven communes (Cotonou, Porto-Novo, Aguégués, Ifangni, Pobè Athiémé, and Grand-Popo) of Southern Benin. Anopheles were collected using indoor/outdoor Human Landing Catches (HLCs) and Pyrethrum Spray Catches (PSCs). Following morphological identification, PCR was used to identify An. gambiae sensu lato (s.l.) to species level and to screen for the presence of both Wolbachia and Microsporidia. Plasmodium falciparum sporozoite infection was also assessed using ELISA.
Results
Overall, species composition in An. gambiae s.l. was 53.7% An. coluzzii, while the remainder was An. gambiae sensu stricto (s.s.). Combined data of the two sampling techniques revealed a mean infection prevalence with Wolbachia of 5.1% (95% CI 0.90–18.6) and 1.3% (95% CI 0.07–7.8) in An. gambiae s.s. and An. coluzzii, respectively. The mean infection prevalence with Microsporidia was 41.0% (95% CI 25.9–57.8) for An. gambiae s.s. and 57.0% (95% CI 45.4–67.9) for An. coluzzii. Wolbachia was only observed in Ifangni, Pobè, and Cotonou, while Microsporidia was detected in all study communes. Aggregated data for HLCs and PSCs showed a sporozoite rate (SR) of 0.80% (95% CI 0.09–2.87) and 0.69% (95% CI 0.09–2.87) for An. gambiae and An. coluzzii, respectively, with a mean of 0.74% (95% CI 0.20–1.90). Of the four individual mosquitoes which harboured P. falciparum, none were also infected with Wolbachia and one contained Microsporidia.
Conclusions
The present study is the first report of natural infections of field-collected An. gambiae s.l. populations from Benin with Wolbachia and Microsporidia. Sustained efforts should be made to widen the spectrum of bacteria identified in mosquitoes, with the potential to develop endosymbiont-based control tools; such interventions could be the game-changer in the control of malaria and arboviral disease transmission.
Similar content being viewed by others
Background
Malaria is an infectious disease caused by a parasite of the Plasmodium genus with half of the global population at risk of this disease. In 2020, there were globally 247 million cases, and 619,000 deaths due to malaria [1]. Sub-Saharan Africa, where Plasmodium falciparum remains the most prevalent malaria parasite, bears the greatest global burden of disease [2]. The cornerstones of malaria vector control have been long-lasting insecticidal nets (LLINs) and indoor residual spraying (IRS) and have averted 1.5 billion malaria cases and 7.6 million malaria deaths, with these interventions accounting for 68% and 10% of these achievements, respectively [3]. With the scale-up of these interventions, the disease burden in Africa is expected to be significantly reduced by 2030. However, widespread insecticide resistance [4] and changes in vector behavior [5] may sabotage elimination in the upcoming decades. For that, biological control tools such as exploitation of Wolbachia, Spiroplasma, and Microsporidia endosymbionts that can be used alone or in combination with insecticide based-tools, have been developed to improve the control of vector-borne diseases, including malaria [6,7,8,9]. These bacteria have a large array of interactions including mutualism, commensalism, and parasitism within their hosts [10]. Wolbachia can colonize certain mosquito populations, and impact pathogen development, thereby reducing their infection and transmission potential [7, 8, 11]. Laboratory experiments have shown an absence of dengue virus infection in populations of Aedes aegypti artificially infected with Wolbachia [7, 12]. In addition, other laboratory trials showed that some Wolbachia strains impede infection of Anopheles vectors with Plasmodium species [13,14,15,16], making it an alternative option for malaria control. However, evidence of an impact of Wolbachia infection on malaria transmission at the community level is still scarce [13, 14]. It has long been assumed that Wolbachia is absent from natural populations of Anopheles [17]. It is only recently that studies have reported that Anopheles gambiae sensu stricto (s.s.), Anopheles coluzzii and Anopheles arabiensis can be found naturally infected by Wolbachia in Burkina Faso and Mali [18,19,20] and Anopheles moucheti and Anopheles demeilloni have been reported infected by Wolbachia in Cameroon, Kenya and the Democratic Republic of the Congo, with evidence of the capacity to induce cytoplasmic incompatibility [15]. Negative correlations between the presence of Wolbachia and development of Plasmodium has been demonstrated in An. gambiae in Mali and An. coluzzii in Burkina Faso [20, 21]. This supports the need for developing new vector control tools based on Wolbachia-Anopheles interactions.
The first report of Microsporidia in An. arabiensis was in Kenya, where Microsporidia infected mosquitoes were unable to be infected with P. falciparum [22]. The presence of this endosymbiont in wild vector populations, warrants screening for it in other endemic regions in Africa.
To progress the development of endosymbiont-based malaria control tools, it is important to continue identifying, and characterizing the native range of endosymbiont-infected Anopheles vector populations. The present study conducted in Southern Benin aims to identify the natural presence of Wolbachia and Microsporidia in Anopheles gambiae s.l., the main malaria vector in this region.
Methods
Study area
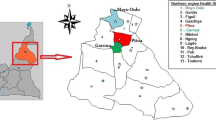
The present study was conducted in September–October 2022 in seven communes (Cotonou, Porto-Novo, Aguégués, Ifangni, Pobè, Athiémé, and Grand-Popo) of Southern Benin (Fig. 1), characterized by a subequatorial climate with two wet (April to July, and September to October), and two dry (November to March, and July to August) seasons. The highest temperatures in the area were between 28 °C and 32 °C, and the lowest between 23 °C and 26 °C. The annual rainfall in the area was approximately 1245 mm, and the main malaria vector species were An. coluzzii and An. gambiae s.s. [23].
Mosquito collections
The present study occurred in September–October 2022. In each study commune, adult mosquitoes were collected using human landing catches (HLCs). In each of two randomly selected houses, two (one indoors and one outdoors) trained collectors were positioned between 08:00 p.m. and 01:00 a.m. and replaced by two others between 01:00 a.m. and 06:00 a.m. Using mouth aspirators and flashlights, they collected all mosquitoes that attempted to bite their lower limbs.
In addition, collection of mosquitoes was also performed using pyrethrum spray catches (PSCs), performed early in the morning in 10 houses selected at random in each surveyed commune. This collection technique consisted of laying white sheets on the floor, closing all openings in the rooms, and spraying aerosol insecticides indoors. After 10–15 min, all indoor resting mosquitoes that fell on the sheets, after insecticidal exposure, were collected using forceps and petri-dishes.
Adult mosquitoes collected through the two sampling techniques were morphologically identified using a binocular loupe, according to the taxonomic keys of Gillies and Coetzee [24], and individually stored on silicagel at − 20 °C for further molecular analyses.
Molecular analyses
Detection of P. falciparum sporozoite infection and molecular species identification
All specimens of An. gambiae sensu lato (s.l.) collected with HLCs and PSCs were analysed using ELISA-CSP to identify P. falciparum sporozoite infection [25]. Molecular species identification was performed in all caught specimens of An. gambiae s.l. using the protocol of Santolamazza et al. [26].
Identification of the presence of endosymbiont
Overall, a total of 118 pools, each containing 5 specimens of either An. gambiae or An. coluzzii were formed. The genomic DNA of these pools was extracted using DNeasy Blood and Tissue kits (Qiagen, France), following the manufacturer instructions:
-
Microsporidia
The following primers: MB18SF (CGCCGGCCGTGAAAAATTTA) and MB18SR (CCTTGGACGTGGGGAGCTATC) were used to detect Microsporidia in An. gambiae s.l. [22]. Each PCR reaction consisted of a final volume of 12.5 µl with 120 ng/µl of DNA, 1 X Hot Start Taq (Thermo Scientific), and 0.3 µM of each primer. The conditions used were an initial denaturation at 95 °C for 5 min, 35 denaturation cycles at 95 °C for 1 min, hybridization at 62 °C for 90 s and an extension at 72 °C for a further 60 s. The final elongation was carried out at 72 °C for 5 min.
-
Molecular detection of Wolbachia
120 ng/µl of DNA was used to amplify a region of the 16S rDNA of Wolbachia using a nested PCR approach, which is specific for natural Wolbachia Anga infections in An. gambiae s.l. [21]. The primer pairs specific to Wolbachia Anga were Forward: 5ʹ-CATACCTATTCGAAGGGATAG-3ʹ; and Reverse: 5ʹ-AGCTTCGAGTGAAACCAATTC-3ʹ [27], which were used for the first reaction. The conditions for this amplification were: 5 min at 95 °C, followed by 45 cycles of 45 s at 95 °C, 45 s at 60 °C, 1 min at 72 °C, and 5 min at 72 °C. This was followed by a second amplification step using 0.3 µM of each primer—Forward: 5′-GAAGGGATAGGGTCGGTCG-3′ and Reverse: 5′-CAATTCCCATGCGTGGACG-3′ in a final reaction volume of 15.5 µl composed of 120 ng/µl of DNA and 1 × Hot Start Taq buffer (Thermo Scientific), using the following conditions: 15 min at 95 °C, followed by 35 cycles of 15 s at 95 °C, 25 s at 66 °C, 1 min at 72 °C, and 5 min at 72 °C [20]. Amplified fragments of 412 bp corresponding to Wolbachia Anga were confirmed by electrophoresis on 2% agarose gels.
Results
Mosquito species composition
Overall, a total of 6225 mosquitoes were collected using HLCs, with a higher ratio (77.8%, n = 4841) outdoors. Indoors, the most frequent mosquito species were Culex quinquefasciatus (57.4%), followed by Mansonia africana (19.7%), and An. gambiae s.l. (15.8%). The same trend was observed outdoors. Other mosquito species such as Anopheles funestus, Ae. aegypti, and other Culex spp. were also collected but at lower frequencies (< 4%) (Fig. 2).
The same trend was observed with PSCs that collected 305 mosquitoes, with Cx. quinquefasciatus being the most frequent mosquito species (64.6%), followed by Mansonia africana (14.8%), and An. gambiae s.l. (9.2%) (Additional file 1: Table S1).
Of 538 specimens of An. gambiae s.l. collected through the two sampling techniques and molecularly speciated, 53.7% (n = 289) were An. coluzzii, while the rest was An. gambiae s.s.
Overall, the predominant species was An. coluzzii in Cotonou (100%), and Athiémé (80.4%), while it was An. gambiae s.s. in Porto-Novo, Aguégués, Ifangni, Pobè, and Grand-Popo with relative frequencies ranging between 66.7–100% (Fig. 3).
Infection prevalence with Wolbachia and Microsporidia
Overall, both Wolbachia (Fig. 4) and Microsporidia (Fig. 5) were identified in An. gambiae s.l. The infection prevalence with Wolbachia was 5.1% (95% CI 0.90–18.6) in An. gambiae s.s. versus 1.3% (95% CI 0.07–7.8) in An. coluzzii (p = 0.53), with a mean of 2.5% (95% CI 0.5–7.3) in the overall species complex (Table 1). Commune level data revealed the presence of Wolbachia in Ifangni, Pobè, and Cotonou (Additional file 1: Table S2).
Infection prevalence with Microsporidia of 41.0% (95% CI 25.9–57.8) in An. gambiae s.s. versus 57.0% (95% CI 45.4–67.9) in An. coluzzii (p = 0.15), with a mean of 53.4% (95% CI 43.9–62.6) in the overall species complex was observed (Table 1). Irrespective of the molecular species, infection to Microsporidia was observed in all study communes (Additional file 1: Table S2).
Sporozoite rate (SR) in An. gambiae s.l. and its molecular species
Of the 538 specimens of An. gambiae s.l. collected, 4 were infected (two from Cotonou, one from Porto-Novo and one from Aguégués), which equated to a mean SR of 0.74% (95% CI 0.20–1.90) (Table 2). At the molecular species level, the SR was 0.80% (95% CI 0.09–2.87) in An. gambiae s.s. vs 0.69% (95% CI 0.08–2.47) in An. coluzzii (p = 1). Of the four individual mosquitoes (two An. coluzzii and two An. gambiae s.s.), which harboured P. falciparum, none was infected with Wolbachia and one contained Microsporidia (An. coluzzii).
The SR of each molecular species observed per study commune is detailed in Additional file 1: Table S3.
Discussion
Given that the efficacy of insecticide-based control tools are under threat because of the emergence of resistance, there is a growing interest in the use of alternative, effective biological vector control strategies. For that, the search for natural endosymbiont-Anopheles systems capable of reducing vector competence has become essential. The present study is the first that reports the presence of Wolbachia and Microsporidia in both An. gambiae s.s. and An. coluzzii in Benin.
A trial recently conducted in Kenya showed that Microsporidia, a vertically transmitted bacteria was capable of disrupting Plasmodium development in An. arabiensis [22]. Moreover, it has been demonstrated that some mosquitoes can have their longevity reduced by Wolbachia, which prevents the completion of the life cycle of some infectious pathogens, thereby interrupting transmission [28]. Findings of the present study shows the natural presence of Microsporidia and Wolbachia in the microbiota of An. gambiae s.l. in Benin. These results confirm those of Gomes et al. [21] and Dada et al. [29] that demonstrated the ability of An. gambiae s.l. to host Wolbachia and Microsporidia.
In the present study, the infection rates (5.1% in An. gambiae s.s. and 1.3% in An. coluzzii) to Wolbachia was overall lower compared to those observed in Burkina-Faso (46% in An. coluzzii and 33% in An. arabiensis). The same trend was observed at the complex level (An. gambiae s.l.), with infection rates ranging between 46 and 78%, depending on the study site in Mali [21]. The general low infection prevalence of Wolbachia in the study area could be due to low density levels that were difficult to detect by PCR or reflect the insensitivity of the end-point PCR technique used. In a previous study in Mali, nested PCR failed to identify 21.7% of infected An. gambiae s.l. samples infected with Wolbachia wAnga-Mali with poor concordance between technical replicates, suggesting that Wolbachia levels were close to the limit of detection of these assays [21]. qPCR methodologies, recently developed for Wolbachia Anga, may have improved detection levels [21]; however, were not feasible with the limited laboratory resources. Thus, the null infection rate to Wolbachia observed in some study communes should not necessarily be interpreted as an absence of this endosymbiont. Taken together these results suggest that natural infection of An. gambiae s.l. to Wolbachia is highly variable across sites in Africa. A similar result was observed in China where the prevalence of Wolbachia natural infection was highly variable in field-collected mosquitoes (Aedes albopictus, Anopheles sinensis, Armigeres subalbatus, Cx pipiens, and Culex tritaeniorhynchus) collected across 25 surveyed provinces [30]. Moreover, Wolbachia natural infection could also be highly variable in various Anopheles species as previously reported in Gabon, Central Africa [31]. Of note, there is a huge diversity of Wolbachia strains with different effects in nature [19].
The deployment of a Wolbachia-based control tool for controlling mosquito borne diseases through the production of sterile insects or pathogen blocking, requires the inducement of cytoplasmic incompatibility to drive the bacterium into natural arthropod populations [32]. While Wolbachia has been shown to impact P. falciparum development, previous works revealed that Wolbachia detected in the present study do not confer cytoplasmic incompatibility [20] and, therefore, would not be feasible to use for control purposes.
The findings show a strong presence of Microsporidia in both An. gambiae s.s. and An. coluzzii, with a mean infection rate of 53.4%. This corroborates previous findings from Akorli et al. [33] who demonstrated that Microsporidia was highly associated with An. gambiae s.s. and An. coluzzii in Ghana. Also, a higher respective, albeit non-significant infection rate to Microsporidia was observed in An. coluzzii than in An. gambiae both in the present trial (57% vs 41%) and the one (80.7% vs 76.0%) of Akorli et al. [34]. Thus, one aspect worth investigating in future trials would be whether An. coluzzii is more susceptible to infection with Microsporidia, compared to An. gambiae s.s.
Though the present trial is a cross-sectional one, it is worth mentioning that investigating the dynamics or variations in bacterial diversity in field-collected adult populations of An. gambiae s.l. is challenging, as bacterial diversity is strongly influenced by several factors such as seasonality, locality-dependent acquisition of environmental microbes [34], diet at larval stage [35], sugar/blood feeding, mating [36], and other factors likely not yet studied.
Overall, in both HLCs and PSCs, the most frequent mosquito species collected were Culex spp, and Mansonia spp, followed by Anopheles spp, and Aedes spp. The same trend was previously observed in Cove, Ouinhi and Zangnanando communes located 156 km away from Cotonou, the economic capital of Benin [37]. Molecular species identification revealed the presence of a mixture of An. coluzzii and An. gambiae s.s. which is consistent with findings from several other previous trials conducted in Southern Benin [21, 35, 36]. Overall, the SR was similar in An. gambiae s.s. and An. coluzzii, which corroborates previous findings from Akogbeto et al. [38] in Northern Benin. However, given the P. falciparum infection rate was assessed at the mosquito level, while the infection rate to each endosymbiont was evaluated at the pool level, it was not possible to assess the influence of the presence of each endosymbiont on the Plasmodium sporozoite infection, which is a limitation for the study. Failure to carry out phylogenetic analyses in order to identify relationships between Microsporidia and Wolbachia detected in An. gambiae s.l. from Benin and those observed in other regions in Africa also constitutes another drawback of this study.
Conclusion
The present study is the first to report the natural presence of both Wolbachia and Microsporidia in natural populations of An. gambiae in Benin. Sustained efforts should be made to widen the spectrum of bacteria identified in mosquitoes, with the potential to develop endosymbiont-based control tools; such interventions could be the game-changer in the control of malaria and arboviral disease transmission.
Availability of data and materials
The data is available on reasonable request from the corresponding author.
References
WHO. World malaria report 2022. Geneva: World Health Organization; 2022. https://www.who.int/teams/global-malariaprogramme/reports/world-malaria-report-2022.
Doumun MS. Contribution au développement des techniques d’analyse d’images multispectrales pour l’aide au diagnostic du paludisme. Université de Lille ; Institut National Polytechnique Félix Houphouët-Boigny (Yamoussoukro, Côte d’Ivoire); 2021.
Bhatt S, Weiss DJ, Cameron E, Bisanzio D, Mappin B, Dalrymple U, et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 2015;526:207–11.
Ranson H, Lissenden N. Insecticide resistance in African Anopheles mosquitoes: a worsening situation that needs urgent action to maintain malaria control. Trends Parasitol. 2016;32:187–96.
Jamet H, Curtis C. Mosquito behavior and vector control. Annu Rev Entomol. 2005;50:53–70.
Bourtzis K, Dobson SL, Xi Z, Rasgon JL, Calvitti M, Moreira LA, et al. Harnessing mosquito-Wolbachia symbiosis for vector and disease control. Acta Trop. 2014;132(Suppl):S150-163.
Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, Muzzi F, et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476:454–7.
McGraw EA, O’Neill SL. Beyond insecticides: new thinking on an ancient problem. Nat Rev Microbiol. 2013;11:181–93.
Iturbe-Ormaetxe I, Walker T, O’Neill SL. Wolbachia and the biological control of mosquito-borne disease. EMBO Rep. 2011;12:508–18.
Werren JH, Windsor D, Guo LR. Distribution of Wolbachia among neotropical arthropods. Proc Biol Sci. 1997;262:197–204.
Hughes GL, Dodson BL, Johnson RM, Murdock CC, Tsujimoto H, Suzuki Y, et al. Native microbiome impedes vertical transmission of Wolbachia in Anopheles mosquitoes. Proc Natl Acad Sci USA. 2014;111:12498–503.
Schmidt TL, Barton NH, Rašić G, Turley AP, Montgomery BL, Iturbe-Ormaetxe I, et al. Local introduction and heterogeneous spatial spread of dengue-suppressing Wolbachia through an urban population of Aedes aegypti. PLoS Biol. 2017;15: e2001894.
Bian G, Joshi D, Dong Y, Lu P, Zhou G, Pan X, et al. Wolbachia Invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science. 2013;340:748–51.
Hughes GL, Koga R, Xue P, Fukatsu T, Rasgon JL. Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathog. 2011;7: e1002043.
Walker T, Quek S, Jeffries CL, Bandibabone J, Dhokiya V, Bamou R, et al. Stable high-density and maternally inherited Wolbachia infections in Anopheles moucheti and Anopheles demeilloni mosquitoes. Curr Biol. 2021;31:2310-2320.e5.
Kambris Z, Blagborough AM, Pinto SB, Blagrove MSC, Godfray HCJ, Sinden RE, et al. Wolbachia stimulates immune gene expression and inhibits Plasmodium development in Anopheles gambiae. PLoS Pathog. 2010;6: e1001143.
Dodson BL, Hughes GL, Paul O, Matacchiero AC, Kramer LD, Rasgon JL. Wolbachia enhances West Nile Virus (WNV) infection in the mosquito Culex tarsalis. PLoS Negl Trop Dis. 2014;8: e2965.
Baldini F, Segata N, Pompon J, Marcenac P, Robert Shaw W, Dabiré RK, et al. Evidence of natural Wolbachia infections in field populations of Anopheles gambiae. Nat Commun. 2014;5:3985.
Hoffmann AA, Ross PA, Rašić G. Wolbachia strains for disease control: ecological and evolutionary considerations. Evol Appl. 2015;8:751–68.
Shaw WR, Marcenac P, Childs LM, Buckee CO, Baldini F, Sawadogo SP, et al. Wolbachia infections in natural Anopheles populations affect egg laying and negatively correlate with Plasmodium development. Nat Commun. 2016;7:11772.
Gomes FM, Hixson BL, Tyner MDW, Ramirez JL, Canepa GE, Alves e Silva TL, et al. Effect of naturally occurring Wolbachia in Anopheles gambiae s.l. mosquitoes from Mali on Plasmodium falciparum malaria transmission. Proc Natl Acad Sci USA. 2017;114:12566–71.
Herren JK, Mbaisi L, Mararo E, Makhulu EE, Mobegi VA, Butungi H, et al. A microsporidian impairs Plasmodium falciparum transmission in Anopheles arabiensis mosquitoes. Nat Commun. 2020;11:2187.
Ossè RA, Tokponnon F, Padonou GG, Glitho ME, Sidick A, Fassinou A, et al. Evidence of transmission of Plasmodium vivax 210 and Plasmodium vivax 247 by Anopheles gambiae and An. coluzzii, major malaria vectors in Benin/West Africa. Insects. 2023;14:231.
Gillies MT, Coetzee M. A supplement to the Anophelinae of Africa south of the Sahara (Afrotropical Region). Publ S Afr Inst Med Res. 1987;55:1–143.
Burkot TR, Graves PM, Cattan JA, Wirtz RA, Gibson FD. The efficiency of sporozoite transmission in the human malarias, P. falciparum and P. vivax. Bull World Health Organ. 1987;65:375–80.
Santolamazza F, Mancini E, Simard F, Qi Y, Tu Z, della Torre A. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar J. 2008;7:163.
Werren JH, Windsor DM. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc Biol Sci. 2000;267:1277–85.
Li Y, Sun Y, Zou J, Zhong D, Liu R, Zhu C, et al. Characterizing the Wolbachia infection in field-collected Culicidae mosquitoes from Hainan Province, China. Parasit Vectors. 2023;16:128.
Dada N, Jupatanakul N, Minard G, Short SM, Akorli J, Villegas LM. Considerations for mosquito microbiome research from the Mosquito Microbiome Consortium. Microbiome. 2021;9:36.
Yang Y, He Y, Zhu G, Zhang J, Gong Z, Huang S, et al. Prevalence and molecular characterization of Wolbachia in field-collected Aedes albopictus, Anopheles sinensis, Armigeres subalbatus, Culex pipiens and Culex tritaeniorhynchus in China. PLoS Negl Trop Dis. 2021;15: e0009911.
Ayala D, Akone-Ella O, Rahola N, Kengne P, Ngangue MF, Mezeme F, et al. Natural Wolbachia infections are common in the major malaria vectors in Central Africa. Evol Appl. 2019;12:1583–94.
Sicard M, Bonneau M, Weill M. Wolbachia prevalence, diversity, and ability to induce cytoplasmic incompatibility in mosquitoes. Curr Opin Insect Sci. 2019;34:12–20.
Akorli J, Akorli EA, Tetteh SNA, Amlalo GK, Opoku M, Pwalia R, et al. Microsporidia MB is found predominantly associated with Anopheles gambiae s.s and Anopheles coluzzii in Ghana. Sci Rep. 2021;11:18658.
Akorli J, Gendrin M, Pels NAP, Yeboah-Manu D, Christophides GK, Wilson MD. Seasonality and locality affect the diversity of Anopheles gambiae and Anopheles coluzzii midgut microbiota from Ghana. PLoS ONE. 2016;11: e0157529.
Saab SA, Dohna H, Nilsson LKJ, Onorati P, Nakhleh J, Terenius O, et al. The environment and species affect gut bacteria composition in laboratory co-cultured Anopheles gambiae and Aedes albopictus mosquitoes. Sci Rep. 2020;10:3352.
Wang Y, Gilbreath TM, Kukutla P, Yan G, Xu J. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS ONE. 2011;6: e24767.
Yovogan B, Sovi A, Padonou GG, Adoha CJ, Akinro B, Chitou S, et al. Pre-intervention characteristics of the mosquito species in Benin in preparation for a randomized controlled trial assessing the efficacy of dual active-ingredient long-lasting insecticidal nets for controlling insecticide-resistant malaria vectors. PLoS ONE. 2021;16: e0251742.
Akogbéto MC, Salako AS, Dagnon F, Aïkpon R, Kouletio M, Sovi A, et al. Blood feeding behaviour comparison and contribution of Anopheles coluzzii and Anopheles gambiae, two sibling species living in sympatry, to malaria transmission in Alibori and Donga region, northern Benin, West Africa. Malar J. 2018;17:307.
Acknowledgements
We are grateful to the people of the surveyed communes as well as their leaders, for their collaboration. We acknowledge field and laboratory technicians from Centre de Recherche Entomologique de Cotonou (CREC) for their dedication and commitment during this work. We also thank Constantin J. Adoha for drawing the map of the study area.
Funding
This study did not received funding from any sources or agency.
Author information
Authors and Affiliations
Contributions
MJA, ASO, LAM, RAO conceived the study; MJA, SH, WS, CZK, SC and BA designed the study; MJA, LT, SH, SC analysed the data; CZK, WS, MJA, ASI, CZK, LT, BA, FT collected the field entomological data; ASO, ASI, FT, GGP, MA, LAM, and RAO collected the molecular data; ASO, FT, GGP, MA, LAM, and RAO supervised the work; MJA and ASO wrote the original draft; MJA, ASO, FT, GGP, MA, LAM, RAO, and GGP critically revised the manuscript for intellectual content. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The protocol of this study was reviewed and approved by the Comité Institutionnel d’Ethique pour la Recherche en Santé du Centre de Recherche Entomologique de Cotonou (CIERS-CREC) (Ethical approval No. 08-22/CREC/CIERS-CREC/SG). Written consent to participate was sought from the heads of the surveyed households and mosquito collectors prior to their involvement. The collectors were trained to sample mosquitoes before they bit. They were vaccinated against yellow fever and regularly checked-up at the nearest health facility. In the occurrence of fever, they were immediately taken care of.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Table S1. Mosquito species composition (PSC data). Table S2. Infection prevalence with Wolbachia Anga and Microsporidia MB per molecular species (An. gambiae s.s. and An. coluzzii) in each study commune (HLC + PSC data). Table S3. SR per molecular species, in each study commune (HLC + PSC data).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ahouandjinou, M.J., Sovi, A., Sidick, A. et al. First report of natural infection of Anopheles gambiae s.s. and Anopheles coluzzii by Wolbachia and Microsporidia in Benin: a cross-sectional study. Malar J 23, 72 (2024). https://doi.org/10.1186/s12936-024-04906-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-024-04906-1