Abstract
Background
Malaria remains a significant cause of morbidity and mortality in Ethiopia with an estimated 3.8 million cases in 2021 and 61% of the population living in areas at risk of malaria transmission. Throughout the country Plasmodium vivax and Plasmodium falciparum are co-endemic, and Duffy expression is highly heterogeneous. The public health significance of Duffy negativity in relation to P. vivax malaria in Ethiopia, however, remains unclear. This study seeks to explore the prevalence and rates of P. vivax malaria infection across Duffy phenotypes in clinical and community settings.
Methods
A total of 9580 and 4667 subjects from community and health facilities from a malaria endemic site and an epidemic-prone site in western Ethiopia were enrolled and examined for P. vivax infection and Duffy expression from February 2018 to April 2021. Association between Duffy expression, P. vivax and P. falciparum infections were examined for samples collected from asymptomatic community volunteers and symptomatic subjects from health centres.
Results
Infection rate of P. vivax among Duffy positives was 2–22 fold higher than Duffy negatives in asymptomatic volunteers from the community. Parasite positivity rate was 10–50 fold higher in Duffy positives than Duffy negatives among samples collected from febrile patients attending health centres and mixed P. vivax and P. falciparum infections were significantly more common than P. vivax mono infections among Duffy negative individuals. Plasmodium vivax parasitaemia measured by 18sRNA parasite gene copy number was similar between Duffy positives and Duffy negatives.
Conclusions
Duffy negativity does not offer complete protection against infection by P. vivax, and cases of P. vivax in Duffy negatives are widespread in Ethiopia, being found in asymptomatic volunteers from communities and in febrile patients from health centres. These findings offer evidence for consideration when developing control and intervention strategies in areas of endemic P. vivax and Duffy heterogeneity.
Similar content being viewed by others
Background
In spite of significant progress towards malaria control in the past two decades, malaria remains a major cause of mortality and morbidity in Africa [1]. According to the World Health Organization, Plasmodium vivax and Plasmodium falciparum contributed to approximately 700 thousand and 230 million cases, respectively, in Africa in 2021 [2]. In Ethiopia, there were an estimated 3.8 million cases in 2021 and 61% of the population resides in areas with endemic transmission [2, 3]. Plasmodium vivax and P. falciparum account for approximately 33% and 67% of all malaria cases, respectively, and it is one of only a few countries in Africa, where P. vivax remains consistently endemic [4].
Current endemicity of P. vivax in Africa correlates with areas of high heterogeneity in Duffy expression [4, 5]. The Duffy antigen receptor for chemokines (DARC), often referred to as the Fy glycoprotein, is a silent heptahelical chemokine receptor located on chromosome 1 and expressed on the surface of erythrocytes. DARC has been recognized as the binding antigen of P. vivax, and a single point mutation located in the GATA-1 transcription factor binding site of the DARC gene promoter (− 67 T > C) causes this receptor to not be expressed, resulting in a Duffy negative phenotype [6, 7]. The absence of this receptor on red blood cells has been shown to confer resistance to blood-stage infection by P. vivax [5, 8, 9]. This negative phenotype is nearly fixed in sub-Saharan Africa, correlating with the general lack of endemic P. vivax on the continent.
Despite this established dogma, cases of P. vivax are being found in confirmed Duffy negative individuals throughout different African countries [10,11,12,13,14]. In addition to Duffy blood group, other population-level factors that influence P. vivax epidemiology include climatic conditions, age, socioeconomic status, access to healthcare, malaria control measures [15, 16]. Whether discovery of increasing number of P. vivax infections in Duffy negatives results from more recent research on P. vivax in Africa or from new P. vivax genetic variants, the data suggest that Duffy negativity no longer confers complete resistance to blood-stage P. vivax infection [17, 18]. Furthermore, Duffy negatives generally develop reduced natural immunity to P. vivax blood-stage antigens [19,20,21].
There remains little information on the public health significance of P. vivax infection in individuals lacking the Duffy antigen in Africa. For example, how frequent are Duffy negative individuals infected with P. vivax compared to Duffy positive individuals from the same communities? How frequently does P. vivax contribute to clinical malaria among Duffy negatives compared to Duffy positives from areas of same endemicities? This population-based study aimed to address these questions in two locations with varying malaria endemicities in southwestern Ethiopia, using samples from communities and health centres.
Methods
Study sites
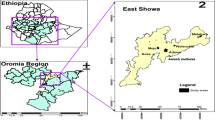
Samples were collected from two study sites, Arjo-Didessa and Gambella, both located in western Ethiopia (Fig. 1) with a rainy season lasting from May to October. The Arjo-Didessa sugarcane plantation is located within the Oromia Regional State 395 km west of the Ethiopian capital Addis Ababa and the area covers most of the Arjo-Didessa sugarcane irrigation scheme. It is located at an elevation ranging from 1200 to 1500 m above sea level, and comprises 15 villages in 3 districts (Jimma Arjo, Bedele District, and Dado Hana District). It contains 1 health centre, 3 health posts, and 9 command posts which are smaller scale health posts located within the temporary residential areas formed by migrant workers. The sugarcane plantation was formerly the Didessa Wildlife Sanctuary before 2006 when the state owned sugarcane plantation was developed to supply the proximal sugarcane factory. It is one of the biggest sugarcane developments in the country, currently covering 5000 hectares with plans to expand to 80,000 hectares [22,23,24]. Gambella is located in the Abobo District in the Gambella Regional State, 811 km west of Addis Ababa. The area’s elevation ranges from 400 to 600 m above sea level and as of 2019 had a population of 20,080. The main socio-economic activity in the area is farming of cotton, maize and sorghum, or working fruit plantations to produce mango, papaya and banana. Additionally, the Alwero Dam provides fishing opportunities and employs approximately 2000 people at a large-scale rice irrigation scheme that currently spans 3000 hectares with plans to expand to 10,000 hectares. The district comprises 19 villages containing 4 health centres and 16 health posts [25, 26]. The populations at both locations primarily consisted of local villagers and migrant workers with long-term residency. These sites were chosen for the study as both areas have high levels of Duffy admixture, and continuous P. vivax endemicity [12, 27].
Map showing location of both study sites; Arjo and Gambella, in western Ethiopia. Includes locations of study clusters, health facilities, and major towns in the regions. The map was created with Esri ArcGIS Pro 3.1 with data sources from field survey, and elevation data from NASA SRTM v3 (https://doi.org/10.5067/MEaSUREs/SRTM/SRTMGL1.003)
Blood sample collection
Finger prick blood samples were collected throughout both study sites from community members who were asymptomatic during cross-sectional surveys, and from febrile volunteers attending health centres from February 2018 to April 2021. From each individual, a total of 3 blood spots, equaling ~ 50 ul, was pressed to Whatman 3MM filter paper for storage and transportation. For community collections all residents willing to participate were included in the study and provided signed informed consent and/or assent for minors under 18 years old. At the time of sample collection, for both clinical and community samples, the age and sex of participants were recorded when possible. Dried blood spots were transported to the University of California Irvine and stored at 4 °C.
DNA extraction and qPCR of Plasmodium species
Parasite DNA was extracted from dried blood spots (DBS) using a standardized saponin/chelex method [28]. DNA was eluted to ~ 200ul molecular grade water stored at 4 °C in the short term or − 20 °C for long term storage. Plasmodium species-specific primers and probes were used to amplify the 18sRNA gene using a previously described protocol with modification [29]. Real time PCR was conducted at a total volume of 12ul containing; 6 µl ThermoFisher FastAdvanced MM (2X), 0.5 µl of each species-specific probe, 0.4 µl of each forward and reverse species-specific primer, and 2 µl parasite DNA. Reaction conditions were set as follows: 50 °C for 2 min, and 45 cycles of 95 °C for 2 min, 95 °C for 3 s, 60 °C for 30 s and run on a QuantStudio 3 Real-Time PCR System.
Duffy sequencing
An approximately ~ 600-bp fragment of the human DARC gene encompassing the − 33rd nucleotide position located in the GATA-1 box of the promoter region was amplified sequenced following established protocols to assess Duffy expression [27, 30, 31]. Specifically, the total volume for amplification was a 20ul reaction mixture containing; 10 µl DreamTaq Green PCR MM (2X), 0.3 µl of each forward and reverse primer, and 2 µl genomic DNA. Thermocycling conditions were set at; 94 °C for 2 min, 35 cycles of 94 °C for 30 s, 61 for 30 s, and 65 for 40 s followed by a 2-min extension at 65 °C. Five microliters of PCR product were run on a 1.5% agarose gel to confirm amplification. PCR product which had successful amplification was cleaned enzymatically to remove remaining primers and dNTPs; 2 µl SAP and 0.2 µl XO1 per was added to PCR product and cleaned via the following thermocycling conditions; 37 °C for 15 min, 80 °C for 15 min, and then held at a 4C extension. Sanger sequencing was conducted by Retrogen Inc. using forward primers and chromatogram results were visually analysed via Chromas for a T → C mutation at the 33rd nucleotide position indicating Duffy negativity. Only samples positive for P. vivax mono and mixed infections were sequenced for Duffy expression.
Data analysis
Malaria prevalence was calculated for both study settings at each study site separately. Overall prevalence of both Plasmodium species was compared between study sites for community and clinical collections via the Chi-Square test for independence. Given that only P. vivax positive samples were sequenced for Duffy expression, rates of Duffy negativity in the population was not directly assessed for this study, the overall rate of P. vivax in Duffy negative and Duffy positive individuals was calculated by dividing the number of P. vivax infections by the expected number of Duffy negative and positive individuals at each site. Expected Duffy negative and positive populations were calculated by multiplying the total number of samples by the Duffy negativity rate in Arjo (43.6%) and Gambella (45.9%) as determined in a previously published study [21]. The ratio of mixed (P. vivax + P. falciparum) to mono (P. vivax) infections was determined for each study setting, community and health facility, for both Duffy negatives and Duffy positives.
Comparisons of the rate of P. vivax in Duffy negatives to Duffy positives, and the ratio of mixed to mono infections for Duffy negatives and positives were made via Fisher’s Exact test for both community and health facility collected samples. Parasite Gene Copy Number (GCN) was calculated from qPCR Cycle threshold (Ct) values via standard curve to estimate parasite density. Log10 transformed GCN was compared between community and health facility settings for both P. vivax and P. falciparum via two-sample t-test, and between Duffy negatives and Duffy positives for both settings via Fisher’s Exact test.
Results
Prevalence of P. vivax across study sites, collection method and Duffy expression
A total of 14,247 dried blood spots were collected from two study sites in southwestern Ethiopia (Fig. 1) from February 2018 to December 2021. Asymptomatic community collections were made via cross-sectional surveys conducted during the spring and late-fall of each year and making up 9580 of the total dried blood spots. The remaining 4667 samples were from symptomatic infections collected from health clinics and facilities in the regions via passive case detection (PCD). In total 344 DBS were positive for only P. vivax, 937 for only P. falciparum and 35 samples exhibited a mixed infection being positive for both P. vivax and P. falciparum (Table 1). A total of 7519 of these DBS were collected from Arjo; 5454 from cross-sectional surveys and 2065 from passive case detection. In Gambella 6728 samples were collected in total; 4126 were collected from the community during cross-sectional surveys and 2602 via passive case detection (Table 1). Overall, P. vivax and P. falciparum infection rate was significantly higher in Gambella than in Arjo (P < 0.001 for both species).
Duffy genotyping was performed only on P. vivax mono infections and mixed P. vivax and P. falciparum infections across all study sites and collection methods (Table 2). Of the 379 P. vivax positive and mixed-species infections, 345 were successfully sequenced at the T33C promoter of the GATA-1 transcription factor. Among the community-based cross-sectional samples, infection rate of P. vivax among the Duffy negatives and positives was low and similar in Arjo, but significantly higher infection rate was found in Gambella among Duffy positives than Duffy negatives (5.6% vs. 0.26%, P < 0.001; Table 2). Similarly, sample positivity rate was more than 10–50 fold higher in Duffy positive than Duffy negatives in both sites among samples collected from the health centre settings (Table 2), suggesting a much reduced P. vivax burden among Duffy negative people in febrile patients.
Interestingly, a considerably large proportion of malaria infections were mixed species infection. Among the 133 successfully sequenced community-based samples, eight out of 133 (6.0%) malaria infections were mixed species and 26 out of 212 (12.3%) samples were mixed infections from the health centre settings (Table 3). Among the Duffy negatives, P. vivax was found more frequently found in the form of mixed-species infection than mono infections, whereas mono P. vivax infections were far more common in Duffy positives. In the community asymptomatic samples, the ratio of mixed species infection to P. vivax mono infection was 0.5 among Duffy negatives, but this ratio was reduced to 0.05 in Duffy positives (P < 0.05; Table 3). In febrile samples from health centres the ratio of mixed species infection to P. vivax mono infection was 3.5 among Duffy negatives, far greater than the ratio observed in Duffy positive (0.10; P < 0.001). This data strongly suggests that in Duffy negative individuals P. vivax is more frequently found in mixed infections compared to P. vivax only mono infections.
Plasmodium vivax parasitaemia in community and clinical samples and across Duffy expressions
Analyses of qPCR data revealed significant differences in the parasitaemia between cross-sectional samples without clinical symptoms and clinical samples collected during passive case detection from health centres for both P. vivax and P. falciparum. In both P. vivax and P. falciparum infections parasitaemia was significantly higher in samples collected via passive case detection than via cross-sectional survey (P < 0.001, Fig. 2). Symptomatic P. vivax infections showed a geometric mean gene copy number (GCN) of 2.03 parasites/µl, which was significantly higher than the asymptomatic P. vivax infections, which had a geometric mean of 0.94 parasites/µl (P < 0.001, Fig. 2). Similarly, symptomatic P. falciparum infections exhibited a geometric mean of 1.67 parasites/µl, which was significantly higher than the asymptomatic P. falciparum infections which had a mean of 0.90 parasites/µl (P < 0.001). Community Duffy-negative and Duffy-positive samples exhibited a similar parasitaemia, with a GCN of 1.28 and 0.93 parasites/µl, respectively (P > 0.05, Fig. 3). Similarly, PCD Duffy-negative and Duffy positive samples showed a mean GCN of 1.93 and 2.07 parasites/µl, respectively (P > 0.05, Fig. 3). These data do not include four Duffy negative samples as their gene copy numbers fell just outside of our standard curve based cut-off range. Given the substantial differences in sample sizes between Duffy-negatives and Duffy-positives it is possible that the lack of significance observed here is indeed due to a small samples size of Duffy negatives.
Violin plots of the log-transformed malaria parasite gene copy number of samples collected from asymptomatic communities and febrile patients of all ages from health centres in Ethiopia. A Plasmodium vivax, and B P. falciparum by qPCR for individuals of all ages. The central box represents the interquartile range with the median shown as the centre line in the box. ***, P < 0.001 based on Fisher’s exact test
Box plots of the log-transformed Plasmodium vivax parasite gene copy number for Duffy negative and Duffy positive individuals of all ages. A asymptomatic community samples; and B febrile malaria samples from health centres. Box plots represent the interquartile range with the median expressed as the centre line. NS, non-significant based on Fisher’s exact test
Discussion
This study sought to examine P. vivax malaria burden in Duffy negative individuals at two field sites with similar proportion of Duffy negativity, but different malaria endemicities in southwest Ethiopia. Plasmodium vivax posed a significant health burden at both sites, but was far more prevalent in the community in Gambella than in Arjo where infection prevalence was over 50 times higher. In febrile patients P. vivax was found more often in Arjo than in Gambella; however, this difference was much less drastic than in the community with Arjo exhibiting only 1.5 times more P. vivax clinical infections than Gambella. Across both sites and collection settings P. vivax was found far less frequently in Duffy negatives than Duffy positives. In the community Duffy positives had approximately 2 and 22-fold greater infection rate of P. vivax than Duffy negatives at Arjo and Gambella, respectively. In febrile patients and samples collected from health facilities this trend was even more apparent; in Arjo and Gambella Duffy positives exhibited a 51 and tenfold greater positivity rate of P. vivax infections, respectively, than Duffy negatives. The variations in rate of infection were highly significant for samples from health centres at both sites, but only significant for community samples from Gambella. The lack of significance in Arjo community samples could potentially be due to the small sample size as only three P. vivax infections were found in the community in Arjo. These strongly suggest that P. vivax infections, though commonly found in Duffy negative individuals, are still predominantly occurring in Duffy positive people. Despite the significant variations in rate of P. vivax infection between Duffy expressions, significant differences in parasitaemia between Duffy negatives and Duffy positives was not observed in either the community or health centres. Perhaps most interestingly this study highlights a pattern of mixed versus mono infections related to Duffy negativity. The ratio of mixed to mono P. vivax infections among Duffy negatives exhibited a 10 and 35-fold greater ratio than Duffy positives in both the community and clinical settings, respectively. Therefore, for Duffy negatives, P. vivax is predominantly found in mixed infections more than mono infections.
Since the level of P. vivax exposure remained consistent among both Duffy positive and Duffy negative individuals across both study locations, the observed diminished burden of P. vivax in Duffy negative individuals underscores that while Duffy negativity does not confer absolute resistance to P. vivax infection, it does exert a significant inhibitory effect on infection establishment. The mechanism behind P. vivax infections of Duffy negatives remains highly elusive, however, several studies have highlighted potential invasion mechanism adaptations of P. vivax that may circumvent Duffy-based infection inhibition and allow for infection on a lesser scale [17, 32]. One of the most well studied of these potential adaptations is the P. vivax Duffy binding protein 1 (PvDBPI) copy number expansion. Several different studies have clearly shown that PvDBP gene amplification both facilitated binding to alternative lower affinity receptors in Duffy negatives, and also suggested that the binding affinity of DARC with high copies of PvDBP could be much higher than with single-copy PvDBP parasites [17, 33,34,35], providing a potential selective pressure towards gene duplication and thus increased infectivity. Two additional ligands, P. vivax glycosylphosphatidylinositol-anchored micronemal antigen (PvGAMA) and P. vivax merozoite surface protein-1 paralog (PvMSP1P), were recently found capable of binding to both Duffy positive and negative red blood cells, suggesting possible involvement in Duffy-independent invasion pathways [36].
Collectively these findings build on previous work documenting P. vivax infections in Duffy negative individuals in numerous African countries [37, 38] including Cameroon [39], Madagascar [10], Angola and Equatorial Guinea [40], Kenya [41], Ethiopia [4]. These studies are consistent with the current findings and support the conclusion that Duffy negative individuals are not completely resistant to infection by P. vivax, yet still have a greatly reduced prevalence of P. vivax infections compared to Duffy positive individuals. This data shows that regardless of exhibiting no significant variation in parasitaemia between Duffy positives and Duffy negatives, several P. vivax infections from Duffy negatives exhibited relatively high levels of parasitaemia potentially implying that these parasites readily infect and adapt to Duffy negativity, allowing for greater erythrocyte invasion. Despite this, several studies have ample evidence that parasitaemia of P. vivax is greatly reduced in Duffy negatives, supporting the hypothesis that parasite infectivity to the human erythrocyte, though not completely inhibited, is indeed reduced in the absence of the Duffy antigen [42, 43]. Several prior studies have also observed that P. vivax infections within Duffy negative individuals are frequently mixed infections, yet these data are limited in that they are predominantly descriptive and do not explore these mixed infections in detail nor compare their prevalence between Duffy negatives and positives [10, 44, 45]. Thus this current study stands out in its efforts to systematically evaluate the prevalence of mixed infections in individuals with Duffy negative status as compared to those with Duffy positive status. These findings thus shed light on the noteworthy phenomenon that P. vivax infections in Duffy negatives frequently encompass mixed-species infections, especially when compared to Duffy positives.
It warrants mention that in the present study is limited in that Duffy expression (negative vs. positive) was inferred based on genotype data of the T33C point mutation in the promoter region of the GATA-1 transcription factor binding site of the Duffy antigen receptor for chemokines (DARC) gene, which is known to alter erythroid expression and eliminate Duffy antigen expression on the red blood cell surface [30, 31, 46]. However, the direct antigen expression (phenotype) was not assessed. It is, therefore, possible for a genotypically categorized Duffy negative individual to potentially express Duffy receptors in some quantity, and the P. vivax strains infecting Duffy negatives in this study may be utilizing such an expression in invasion, despite genotypic negativity. Additionally, neither prevalence nor burden of P. falciparum across Duffy negatives and positives as Duffy expression is not known to be associated with P. falciparum infection.
Conclusion
Understanding the distribution of P. vivax in Africa and exploring the significance of Duffy expression continues to be a challenging and intricate endeavour. Given the low parasitaemia often associated with P. vivax infections of Duffy negative individuals, microscopy and RDTs are often not sensitive enough to detect infection, hindering their diagnosis and study in the field. Indeed, corresponding microscopy data from this area accounted for only approximately 70%, of all qPCR confirmed P. vivax positive infections [22], highlighting the need for more sensitive molecular detection tools in the field. This has significant implications for malaria elimination on the continent as a high proportion of P. vivax cases are likely being overlooked by traditional diagnostic methods. This work shows that not only does P. vivax transmission remain widespread in Ethiopia, but these asymptomatic community infections make up a significant portion of P. vivax cases resulting in a large undetected parasite reservoir that may greatly complicate and hinder interventions and elimination efforts. Finally, it is clear through the current study that Duffy negativity is not a definitive barrier to infection, and P. vivax infections were detected in Duffy negative individuals, encompassing both asymptomatic and febrile malaria instances, frequently occurring in mixed infections. Importantly, these trends persist across both study sites representing high and low endemic settings. This information is vital to informing control and elimination strategies in areas of sub-Saharan Africa with variable P. vivax endemicity and of high Duffy heterogeneity.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Girum T, Shumbej T, Shewangizaw M. Burden of malaria in Ethiopia, 2000–2016: findings from the Global Health Estimates 2016. Trop Dis Travel Med Vaccines. 2019;5:11.
WHO. World Malaria Report. Geneva, World Health Organization, 2022.
Ayele DG, Zewotir TT, Mwambi HG. Prevalence and risk factors of malaria in Ethiopia. Malar J. 2012;11:195.
Ketema T, Bacha K, Getahun K, Portillo HAD, Bassat Q. Plasmodium vivax epidemiology in Ethiopia 2000–2020: a systematic review and meta-analysis. PLoS Negl Trop Dis. 2021;15: e0009781.
Twohig KA, Pfeffer DA, Baird JK, Price RN, Zimmerman PA, Hay SI, et al. Growing evidence of Plasmodium vivax across malaria-endemic Africa. PLoS Negl Trop Dis. 2019;13: e0007140.
Langhi DM Jr, Bordin JO. Duffy blood group and malaria. Hematology. 2006;11:389–98.
Höher G, Fiegenbaum M, Almeida S. Molecular basis of the Duffy blood group system. Blood Transfus. 2018;16:93–100.
Miller LH, Mason SJ, Clyde DF, McGinnis MH. The resistance factor to Plasmodium vivax in blacks The Duffy blood group genotype, FyFy. N Engl J Med. 1976;295:302–4.
Howes RE, Patil AP, Piel FB, Nyangiri OA, Kabaria CW, Gething PW, et al. The global distribution of the Duffy blood group. Nat Commun. 2011;2:266.
Ménard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A, et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci USA. 2010;107:5967–71.
Wurtz NL, Mint LK, Bogreau H, Pradines B, Rogier C, Boukhary MS, et al. Vivax malaria in Maritania includes infection of a Duffy-negative individual. Malar J. 2011;10:336.
Woldearegai TG, Kremsner PG, Kun JF, Mordmüller B. Plasmodium vivax malaria in Duffy-negative individuals from Ethiopia. Trans R Soc Trop Med Hyg. 2013;107:328–31.
Abdelraheem MH, Albsheer MM, Mohamed HS, Amin M, Mahdi Abdel Hamid M. Transmission of Plasmodium vivax in Duffy-negative individuals in central Sudan. Trans R Soc Trop Med Hyg. 2016;110:258–60.
Russo G, Faggioni G, Paganotti GM, Djeunang Dongho GB, Pomponi A, De Santis R, et al. Molecular evidence of Plasmodium vivax infection in Duffy negative symptomatic individuals from Dschang, West Cameroon. Malar J. 2017;16:74.
Habtamu K, Petros B, Yan G. Plasmodium vivax: the potential obstacles it presents to malaria elimination and eradication. Trop Dis Travel Med Vaccines. 2022;8:27.
Auburn S, Cheng Q, Marfurt J, Price RN. The changing epidemiology of Plasmodium vivax: Insights from conventional and novel surveillance tools. PLOS Medicine. 2021;18(4): e1003560. https://doi.org/10.1371/journal.pmed.1003560
Gunalan K, Niangaly A, Thera MA, Doumbo OK, Miller LH. Plasmodium vivax infections of Duffy-negative erythrocytes: historically undetected or a recent adaptation? Trends Parasitol. 2018;34:420–9.
Wilairatana P, Masangkay FR, Kotepui KU, De Jesus MG, Kotepui M. Prevalence and risk of Plasmodium vivax infection among Duffy-negative individuals: a systematic review and meta-analysis. Sci Rep. 2022;12:3998.
Kano FS, de Souza AM, de Menezes TL, Costa MA, Souza-Silva FA, Sanchez BAM, et al. Susceptibility to Plasmodium vivax malaria associated with DARC (Duffy antigen) polymorphisms is influenced by the time of exposure to malaria. Sci Rep. 2018;8:13851.
Maestre A, Muskus C, Duque V, Agudelo O, Liu P, Takagi A, et al. Acquired antibody responses against Plasmodium vivax infection vary with host genotype for duffy antigen receptor for chemokines (DARC). PLoS ONE. 2010;5: e11437.
Bradley L, Yewhalaw D, Hemming-Schroeder E, Embury E, Lee M-C, Zemene E, et al. Determination of Plasmodium vivax and Plasmodium falciparum malaria exposure in two Ethiopian communities and its relationship to Duffy expression. Am J Trop Med Hyg. 2023;109:1028–35.
Hawaria D, Getachew H, Zhou G, Demissew A, Habitamu K, Raya B, et al. Ten years malaria trend at Arjo-Didessa sugar development site and its vicinity, Southwest Ethiopia: a retrospective study. Malar J. 2019;18:145.
Jiang A-L, Lee M-C, Zhou G, Zhong D, Hawaria D, Kibret S, et al. Predicting distribution of malaria vector larval habitats in Ethiopia by integrating distributed hydrologic modeling with remotely sensed data. Sci Rep. 2021;11:10150.
Demissew A, Hawaria D, Kibret S, Animut A, Tsegaye A, Lee MC, et al. Impact of sugarcane irrigation on malaria vector Anopheles mosquito fauna, abundance and seasonality in Arjo-Didessa, Ethiopia. Malar J. 2020;19:344.
Taffese HS, Hemming-Schroeder E, Koepfli C, Tesfaye G, Lee M-C, Kazura J, et al. Malaria epidemiology and interventions in Ethiopia from 2001 to 2016. Infect Dis Poverty. 2018;7:103.
Haileselassie W, Parker DM, Taye B, David RE, Zemene E, Lee M-C, et al. Burden of malaria, impact of interventions and climate variability in Western Ethiopia: an area with large irrigation based farming. BMC Public Health. 2022;22:196.
Lo E, Yewhalaw D, Zhong D, Zemene E, Degefa T, Tushune K, et al. Molecular epidemiology of Plasmodium vivax and Plasmodium falciparum malaria among Duffy-positive and Duffy-negative populations in Ethiopia. Malar J. 2015;14:84.
Bereczy SM, Martensson A, Gil JP, Färnert A. Rapid DNA Extraction from archive blood spots on filter paper for genotyping Plasmodium falciparum. Am J Trop Med Hyg. 2005;72:249–51.
Véron V, Simon S, Carme B. Multiplex real-time PCR detection of P. falciparum, P. vivax and P. malariae in human blood samples. Exp Parasitol. 2009;121:346–51.
King CL, Adams JH, Xianli J, Grimberg BT, McHenry AM, Greenberg LJ, et al. Fy(a)/Fy(b) antigen polymorphism in human erythrocyte Duffy antigen affects susceptibility to Plasmodium vivax malaria. Proc Natl Acad Sci USA. 2011;108:20113–8.
Lo E, Hostetler JB, Yewhalaw D, Pearson RD, Hamid MMA, Gunalan K, et al. Frequent expansion of Plasmodium vivax Duffy binding protein in Ethiopia and its epidemiological significance. PLoS Negl Trop Dis. 2019;13: e0007222.
Ménard D, Chan ER, Benedet C, Ratsimbasoa A, Kim S, Chim P, et al. Whole genome sequencing of field isolates reveals a common duplication of the Duffy binding protein gene in Malagasy Plasmodium vivax strains. PLoS Negl Trop Dis. 2013;7: e2489.
Gunalan K, Lo E, Hostetler JB, Yewhalaw D, Mu J, Neafsey DE, et al. Role of Plasmodium vivax Duffy-binding protein 1 in invasion of Duffy-null Africans. Proc Natl Acad Sci USA. 2016;113:6271–6.
Pearson RD, Amato R, Auburn S, Miotto O, Almagro-Garcia J, Amaratunga C, et al. Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet. 2016;48:959–64.
Hostetler JB, Lo E, Kanjee U, Amaratunga C, Suon S, Sreng S, et al. Independent origin and global distribution of distinct Plasmodium vivax Duffy binding protein gene duplications. PLoS Negl Trop Dis. 2016;10: e0005091.
Popovici J, Roesch C, Rougeron V. The enigmatic mechanisms by which Plasmodium vivax infects Duffy-negative individuals. PLoS Pathog. 2020;16: e1008258.
Zimmerman PA. Plasmodium vivax infection in Duffy-negative people in Africa. Am J Trop Med Hyg. 2017;97:636–8.
Lo E, Russo G, Pestana K, Kepple D, Abagero BR, Dongho GBD, et al. Contrasting epidemiology and genetic variation of Plasmodium vivax infecting Duffy-negative individuals across Africa. Int J Infect Dis. 2021;108:63–71.
Ngassa Mbenda HG, Das A. Molecular evidence of Plasmodium vivax mono and mixed malaria parasite infections in Duffy-negative native Cameroonians. PLoS ONE. 2014;9: e103262.
Mendes C, Dias F, Figueiredo J, Mora VG, Cano J, de Sousa B, et al. Duffy negative antigen is no longer a barrier to Plasmodium vivax–molecular evidences from the African West Coast (Angola and Equatorial Guinea). PLoS Negl Trop Dis. 2011;5: e1192.
Ryan JR, Stoute JA, Amon J, Dunton R, Mtalib R, Koros J, et al. Evidence for transmission of Plasmodium vivax amond a Duffy antigen negative population in Western Kenya. Am J Trop Med Hyg. 2006;74:575–81.
Albsheer MMA, Pestana K, Ahmed S, Elfaki M, Gamil E, Ahmed SM, et al. Distribution of Duffy phenotypes among Plasmodium vivax infections in Sudan. Genes (Basel). 2019;10:437.
Abate A, Bouyssou I, Mabilotte S, et al. Vivax malaria in Duffy-negative patients shows invariably low asexual parasitaemia: implication towards malaria control in Ethiopia. Malar J. 2022;21:230.
Oboh MA, Badiane AS, Ntadom G, Ndiaye YD, Diongue K, Diallo MA, et al. Molecular identification of Plasmodium species responsible for malaria reveals Plasmodium vivax isolates in Duffy negative individuals from southwestern Nigeria. Malar J. 2018;17:439.
Oboh MA, Singh US, Ndiaye D, Badiane AS, Ali NA, Bharti PK, et al. Presence of additional Plasmodium vivax malaria in Duffy negative individuals from Southwestern Nigeria. Malar J. 2020;19:229.
Tournamille C, Colin Y, Cartron JP, Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy–negative individuals. Nat Genet. 1995;10:224–8.
Acknowledgements
The authors sincerely thank the field team at Jimma University in Ethiopia and volunteers for their contribution. We thank the two anonymous reviewers for their valuable comments.
Funding
This study is funded by the National Institutes of Health (5 F31 AI161887, U19 AI129326 and D43 TW001505). These funders were not involved in study design or development, data collection and analysis, publication process or manuscript preparation.
Author information
Authors and Affiliations
Contributions
LB, and GY conceived this paper; LB drafted it with contributions from MCL. LB, EZ, TD and DY provided field data; LB, EHS and BJ conducted laboratory analyses. Findings were reviewed and interpreted by LB and GY with contributions from EL, EHS and DY; EL, CK, JK, GY edited and revised the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical and scientific approval and clearance was obtained from the institutional scientific and ethical review boards of the University of California, Irvine, USA and Jimma University, Ethiopia. Written informed consent/assent for participation in the study was obtained from all participants and/or parents/guardians (for minors under the age of 18).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bradley, L., Yewhalaw, D., Hemming-Schroeder, E. et al. Epidemiology of Plasmodium vivax in Duffy negatives and Duffy positives from community and health centre collections in Ethiopia. Malar J 23, 76 (2024). https://doi.org/10.1186/s12936-024-04895-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-024-04895-1