Abstract
Background
Duffy blood group polymorphisms are important in areas where Plasmodium vivax is present because this surface antigen is thought to act as a key receptor for this parasite. In the present study, Duffy blood group genotyping was performed in febrile uninfected and P. vivax-infected patients living in the city of Nouakchott, Mauritania.
Methods
Plasmodium vivax was identified by real-time PCR. The Duffy blood group genotypes were determined by standard PCR followed by sequencing of the promoter region and exon 2 of the Duffy gene in 277 febrile individuals. Fisher's exact test was performed in order to assess the significance of variables.
Results
In the Moorish population, a high frequency of the FYBES/FYBES genotype was observed in uninfected individuals (27.8%), whereas no P. vivax-infected patient had this genotype. This was followed by a high level of FYA/FYB, FYB/FYB, FYB/FYBES and FYA/FYBES genotype frequencies, both in the P. vivax-infected and uninfected patients. In other ethnic groups (Poular, Soninke, Wolof), only the FYBES/FYBES genotype was found in uninfected patients, whereas the FYA/FYBES genotype was observed in two P. vivax-infected patients. In addition, one patient belonging to the Wolof ethnic group presented the FYBES/FYBES genotype and was infected by P. vivax.
Conclusions
This study presents the Duffy blood group polymorphisms in Nouakchott City and demonstrates that in Mauritania, P. vivax is able to infect Duffy-negative patients. Further studies are necessary to identify the process that enables this Duffy-independent P. vivax invasion of human red blood cells.
Similar content being viewed by others
Background
Malaria remains one of the most important parasitic infections in the world, with almost 225 million cases of infection and 0.78 million deaths in 2009, mainly in Africa, Asia and South America [1]. It is caused by infection with one or more of five species of Plasmodium parasites. Plasmodium vivax is the second most common cause of malaria in the world after Plasmodium falciparum, with more than 80 million clinical cases annually. Unlike P. falciparum, P. vivax rarely causes mortality, but it can potentially lead to severe complications and is thereby responsible for considerable morbidity and economic loss in endemic countries [2–8]. Moreover, P. vivax has a wider geographical range, potentially exposing more people to risk of infection (2.85 billion across three continents) [9–11], and it is more difficult to control because of the hypnozoïte forms of the parasite [12, 13]. The presence of P. vivax in Mauritania was first reported in 1948 [14]. More recently, several studies conducted in Nouakchott, the capital of Mauritania, revealed a high proportion of P. vivax, followed by Plasmodium ovale and P. falciparum; autochthonous malaria cases exist but are relatively uncommon [15–17]. In 2009-2010, the prevalence of P. vivax among malaria in children in Nouakchott represented 97.1% [18].
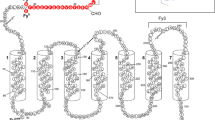
One of the main biological differences between P. vivax and other human malaria parasites is that only P. vivax merozoites use the human Duffy antigen/chemokine receptor (DARC) to invade red blood cells (RBCs) [19–21]. The Duffy antigen was originally identified as a blood group antigen on the surface of RBCs, but it has since been found to be expressed in endothelial cells and neurons [22–24]. It is implicated in multiple chemokine inflammation, inflammatory diseases, and cancer and might play a role in HIV infection [25–27]. The DARC gene (also referred to as FY or Duffy), located on chromosome 1, comprises two exons and produces a protein that has a glycosylated external N-terminal domain, seven transmembrane domains and a short cytosolic C-terminal domain that is not coupled to G-proteins or other known intracellular effectors [28–33].
DARC has two main variant forms, Fya and Fyb antigens, which differ by a single amino acid (Gly42Asp) in the NH2 extracellular domain of the polypeptide and are encoded by the alleles FYA and FYB, respectively, which are differentiated by a single base substitution (G125A) [34–36]. The FYA/FYB frequency shows marked geographic disparities; the FYB allele is highly predominant in Africa, while the FYA allele is dominant in Asia [37]. The Duffy blood group has four major phenotypes: Fy(a+b+), Fy(a+b-), Fy(a-b+) and Fy(a-b-). Duffy expression is disrupted by a T to C substitution in the gene's promoter region at nucleotide -33, preventing the binding of the h-GATA-1 erythroid transcription factor and resulting in the null expression of the Duffy gene in erythroid cells only [38–40]. This variant is commonly associated with the FYB allele (corresponding to the FYBES allele, ES stands for "erythroid silent"), although the same mutation has been detected and associated with the FYA allele in individuals living in P. vivax-endemic region of Papua New Guinea (FYAES ) [41]. The FYBES allele is almost fixed in West and Central Africa, and as a consequence, the Fy(a-b-) (null) phenotype is predominant among populations of West and Central African descent. This phenotype is rare among Caucasian, Amerindian, Indian and Asian populations. The FYAES mutation is rare and so far appears to be present only in the Melanesian and Tunisian population) [41–44]. Other rare variants have been described, most notably the FYX allele, which occurs mainly in Caucasians [45, 46] and is characterized by a weak expression of Fyb antigen (Fybweak).
Some authors [47–50] have attributed the FYX allele to a single polymorphism of the FYB allele (C265T→Arg89Cys) (FYX1), while others have indicated two (C265T and G298A→Ala100Thr) (FYX2) [51–53] or even three polymorphisms (C265T, G298A and G145T→ Ala49Ser) (FYX3) [54]. The point mutation G298A alone did not cause a decrease of the Fyb expression [47]. This allele is also named FYB* in the present study. Eight combinations of alleles (FYA, FYB, FYB*, FYX [FYX1, FYX2 and FYX3], FYAES and FYBES ) result in 32 different genotypes (Additional files 1 and 2).
Malaria therapy, as well as experimental and epidemiological studies, have shown that erythrocyte Duffy blood group negative individuals, mostly of African ancestry, are resistant to P. vivax infection [21]. However, several reports have provided evidence for P. vivax infections among Duffy-negative patients [55–59], suggesting that there are P. vivax strains that have acquired a Duffy-independent mechanism of erythrocyte invasion. Little is known about the frequency of Duffy polymorphisms in Mauritanian populations, especially in P. vivax-infected individuals. The objective of the present study was to evaluate the Duffy blood group allelic and genotype frequencies in the city of Nouakchott and to compare these frequencies between P. vivax- infected and uninfected febrile patients.
Methods
Study populations
This study was conducted in the capital and largest city of Mauritania, Nouakchott, which is located on the Atlantic coast of the Sahara Desert (18°.11'N; 16°.16'W). The city is divided into nine districts and consists of approximately 800,000 inhabitants. Nouakchott features an arid climate with a short wet season extending from July to September. The city has five hospitals and eleven health centres. Between 2007 and 2009, Lekweiry et al conducted a preliminary study on the incidence of malaria in Nouakchott [17].
Capillary blood samples from 439 febrile outpatients from all Nouakchott districts who were seen in the two main hospitals of the city (National Hospital and Chiekh Zayed Hospital) and in the District Health Center of Teyarett were collected onto Whatman 3 MM filter paper. A subset of 277 patients were enrolled in this study to evaluate Duffy blood group. Of these 277 patients, 110 had a positive P. vivax diagnosis and 167 were not infected with Plasmodium but their individual data on the place of residence and/or ethnic group membership were available.
Consent
This study was reviewed and approved by the Mauritanian National Ethics Committee.
Genomic DNA extraction
Blood samples were spotted onto Whatman 3 MM filter paper, dried, and stored at room temperature until use. DNA was extracted with the MagMAX™-96 DNA Multi-Sample Kit according to the manufacturer's instructions using a MagMAX™ Express-96 Magnetic Particle Processor (Applied Biosystems, Courtaboeuf, France).
Identification of Plasmodium species by real-time PCR
Plasmodium detection was performed by real-time LightCycler® PCR (Roche, Meylan, France). The following oligonucleotides primers and probes designed with Primer Express software v2.0 (Applied Biosystems) were used: forward-5'-TTTATGTATTGGTATAACATTCGG-3', reverse-5'-GGCAAATAACTTTATCATAGAATTGAC-3' and probe-5'-FAM- TACACTACCAACACATGGGGCTACAAGAGGT-BBQ-3' for P. falciparum aquaglyceroporin gene (AJ413249); forward-5'-GTGGCCGCCTTTTTGCT-3', reverse-5'-CCTCCCTGAAACAAGTCATCG-3' and probe-5'-HEX- CATCTACGTGGACAACGGGCTCAACA-BHQ1-3' for P. vivax enoyl-acyl carrier protein reductase gene (AY423076); forward- 5'-GAGGAATGGTCACCATGTAGTGT-3', reverse-5'-CAAATTTCAGTTTCAAGGTCACTTAA-3' and probe-5'-HEX- ATTTTTTGCATCAACCTTTCTTCTAGCCC -BHQ1-3' for Plasmodium malariae circumsporozoïte gene (S69014); and forward-5'-CCAAGCCCAGATAATAAGGAAGGT3', reverse-5'-TTCGTGCACTTCAACTTACATTCAGT-3' and probe-5'-FAM-TTATTGTCCTCTGGGTTTGGAACTTTGCC-BBQ-3' for P. ovale P25 ookinete surface protein gene (AB074973) (Eurogentec, Angers, France). Each parasite species was detected separately. Individual PCR amplifications were carried out using 4 μl of 5× concentrate Master Mix (LightCycler® TaqMan® Master, Roche), 0.8 μM of each primer, 0.1 μM of probe and 5 μL of template DNA in a final volume of 20 μL. The thermal cycling conditions were 95°C for 10 min and 45 cycles of 95°C for 10 sec and 60°C for 30 sec, followed by a cooling step of 40°C for 30 sec. For each PCR run, two negative controls (water and human DNA) and a positive control (DNA from each species) were used. Fluorescence acquisition was performed at the end of each extension step.
Duffy blood group genotyping
Duffy blood group genotypes were assessed using PCR amplification of the human Duffy antigen/chemokine receptor gene (NG_011626.1) followed by sequencing. The promoter region that flanks the GATA box motif (a fragment of 392 bp) was amplified using the following primers, which were designed with the NCBI/Primer-BLAST online tool [60]:
forward-5'-CCCAAGGCCAGTGACCCCCATA-3' and reverse-5'-AGAGGGAGCTAGGAGGCTAGCAT-3' (Eurogentec). To determine the Duffy RBC polymorphism, a 541-bp fragment spanning part of intron and exon 2 was amplified using the following primers (also designed with the NCBI/Primer-Blast online tool [60]): forward-5'-CCTGCAGAGACCTTGTTCTCCCAC-3' and reverse-5'-AGCAGCAAAGCCTGGGCAAAGG-3' (Eurogentec).
The reaction mixture for both PCR amplifications included 10 μl of genomic DNA, 2.5 μl of 10× reaction buffer (Eurogentec), 0.5 μM of each primer, 200 μM of deoxynucleoside triphosphate mixture (dGTP, dATP, dTTP and dCTP) (Euromedex, Souffelweyersheim, France), 1.5 mM of MgCl2 and 2.5 units of RedGoldStar® DNA polymerase (Eurogentec) in a final volume of 25 μL. The thermal cycler (T3 Biometra, Archamps, France) was programmed as follows: an initial 94°C incubation for 2 min followed by 40 cycles of 94°C for 30 sec, 58°C for 30 sec and 72°C for 25 sec for the promoter region and 40 cycles of 94°C for 30 sec, 58°C for 30 sec and 72°C for 35 sec for the segment covering part of intron and exon 2. A final 5-min extension step was performed at 72°C for both regions. The PCR products were loaded on 1.5% agarose gel containing 0.5 μg/mL ethidium bromide. Amplicons were purified using the QIAquick 96 PCR BioRobot Kit and an automated protocol on the BioRobot 8000 workstation (Qiagen, Courtaboeuf, France). The purified fragments were sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) using the primers described above. The sequencing reaction products were purified using the BigDye XTerminator® Purification Kit (Applied Biosystems) in accordance with the manufacturer's instructions. The purified products were sequenced using an ABI Prism 3100 analyser (Applied Biosystems). Sequences were analysed using Vector NTI advance™ software (version 11, Invitrogen, Cergy Pontoise, France).
Statistical analysis
Fisher's exact test was used to compare the proportions of Duffy genotypes in relation to P. vivax infection and ethnic origin (GraphPad Prism v5.01). The significance level was fixed at P < 0.05.
Results
Plasmodium species diagnosis
The results obtained by real-time PCR were in accordance with the previous data obtained for species diagnosis (Malaria Rapid Diagnostic Test, microscopy and nested PCR performed by [17]). Of 277 outpatients, 110 were positive for P. vivax. These patients came from various Nouakchott districts (Additional file 3).
Duffy genotypes in febrile uninfected patients and P. vivax-infected patients from Nouakchott
The promoter region and exon 2 of the Duffy gene from each sample selected for the study were amplified and sequenced (Additional file 3). A comparison of Duffy genotypes, phenotypes and allele frequencies according to the ethnic groups between P. vivax-infected and malaria-free patients is presented in Tables 1 and 2. The Moorish population represented the majority of patients (83%). Only a few patients belonged to the other ethnic groups: Poular (4%), Soninke (1%) and Wolof (1%). Information on ethnic origin was not available for some patients (11%), but these patients were included in the study because they were positive for P. vivax. The complete sequence of the Duffy gene was obtained for 258 patients (93%).
In the Moorish population, the prevalence rate of the FYBES/FYBES genotype (Fy(a-b-) phenotype) was 27.8% (n = 40) and 0% among uninfected and P. vivax-infected individuals, respectively (p < 0.0001, Fisher's exact test). This was followed by a high level of FYA/FYB, FYB/FYB, FYB/FYBES and FYA/FYBES genotype frequencies (Fy(a+b+), Fy(a-b+), Fy(a-b+) and Fy(a+b-) phenotypes, respectively) in both P. vivax-infected and uninfected patients. Low frequencies were detected for the FYA/FYA, FYA/FYB*, FYB*/FYBES and FYB/FYB* genotypes (Fy(a+b-), Fy(a+b+), Fy(a-b+) and Fy(a-b+) phenotypes, respectively) in both infected and uninfected patients. In the other ethnic groups (Poular, Soninke and Wolof), only the FYBES/FYBES genotype was found in uninfected patients, whereas the FYA/FYBES genotype was observed in two P. vivax-infected patients, Soninke ethnic.
One P. vivax-infected patient presented the FYBES/FYBES genotype, resulting in a Duffy-negative phenotype. This patient was a two-year-old female, belonging to the Wolof ethnic group and living in the district of Dar Naim.
Discussion
Recent reports on P. vivax infections suggest that this parasite may be evolving and adapting to new epidemiological contexts, becoming not only more virulent but also more frequent in countries where the incidence has traditionally been low [9–12, 61, 62]. The evaluation of Duffy blood group polymorphisms is important in areas where P. vivax prevails, as the Duffy antigen serves as a receptor on the surface of RBCs. Until now, few studies have reported the presence of P. vivax in Mauritania [15, 17, 18, 29, 63] and only one study assessed the distribution of Duffy polymorphisms in Nouakchott [64]. In the present work, the evaluation of Duffy blood group genotypes was undertaken in diverse/multiple human populations that included P. vivax-infected, uninfected, Duffy-positive and Duffy-negative people to i) assess the Duffy gene polymorphism within a cosmopolitan African community and ii) determine whether P. vivax is able to penetrate into RBCs in Duffy-negative patients, who have been thought to be resistant to P. vivax infection [21].
The Mauritanian population has a highly heterogeneous ethnic composition. It is primarily constituted of Moors (an ethnicity with a mix of Arab and Berber ancestry) who live in the North of the country and various black ethnic groups, including Soninke, Wolof and Poular, in the South. Duffy gene polymorphism among different ethnic groups is a characteristic of this blood system and has been used as a marker of ethnic composition as well as an indicator of the evolution of human populations. In 1986, Lepers et al undertook the study of Duffy blood group in 107 individuals belonging to different ethnic groups and residing in Nouakchott [64]. In the overall population, 27% of the individuals were Duffy-positive, whereas the others were Duffy-negative. The proportion of Duffy- positive individuals differed according to the ethnic groups: 54% of Moors were Fy+, while only 2% of black ethnic groups were Fy+.
In the current study, slight differences were observed in the global population: 78% of the individuals were Fy+ and 22% were Fy-. The FYA/FYB genotype was the most common, followed by the heterozygotes FYA/FYBES and FYB/FYBES and the homozygous FYB alleles. It should be noted that no patient had the phenotype Fy(a+bweak) or Fy(a-bweak), as the allele FYX was not present in the population.
When compared to other North African populations, the frequencies of FYA and FYB alleles are similar to that observed in the Tunisian people [44, 65], while the allelic frequency of FYBES and the lack of FYX are similar to what is observed in Morocco [66]. Overall, FYA and FYB alleles are mainly represented in Europe, while the allele FYBES is predominant in Africa [37].
The presence of P. vivax in Mauritania was first reported in 1948 [14] and confirmed in two recent studies suggesting autochthonous P. vivax transmission in some patients who had never travelled outside Nouakchott [15, 17]. RBCs of Duffy-negative individuals seem to be naturally resistant to invasion by the P. vivax human malaria parasite [21]. The present study describes for the first time that one Duffy-negative patient living in Nouakchott, i.e., in North Africa, was infected with P. vivax. The identification of P. vivax was performed by real-time PCR, and the Duffy genotypes were determined by sequencing, making it unlikely that a parasite other than P. vivax was involved.
Our data thereby confirmed the suspicion of some authors, who also believe that P. vivax could be evolving to use receptors other than Duffy to invade erythrocytes in patients in Brazil [55, 56], Kenya [59], and more recently, in Madagascar [57], Angola and Equatorial Guinea [58]. As suggested in previous studies [57, 58], Duffy-positive individuals may serve as reservoirs for P. vivax, allowing this parasite to infect hepatocytes of Duffy-negative individuals and select for new P. vivax strains with the capacity to invade Duffy-negative erythrocytes.
Conclusions
Further analyses are needed to understand the dynamics of the Duffy gene and its possible contribution as a modulator in the susceptibility to malaria. The data obtained in the present study emphasize the importance of the evaluation of Duffy blood group genotypes in P. vivax malaria endemic areas. The results of the present study support the hypothesis that Duffy-negative individuals from North Africa could be infected by P. vivax and that this parasite may be rapidly evolving to use other receptors than Duffy to invade the erythrocytes. Further longitudinal studies on P. vivax and host-parasite interactions are required to test the validity of these hypotheses. Furthermore, a better understanding of the alternative pathways used by P. vivax to invade human RBCs should become a research priority.
References
WHO Global Malaria Programme: World Malaria Report. 2010, Geneva: World Health Organization
Anstey NM, Russell B, Yeo TW, Price RN: The pathophysiology of vivax malaria. Trends Parasitol. 2009, 25: 220-227. 10.1016/j.pt.2009.02.003.
Barcus MJ, Basri H, Picarima H, Manyakori C, Sekartuti , Elyazar I, Bangs MJ, Maguire JD, Baird JK: Demographic risk factors for severe and fatal vivax and falciparum malaria among hospital admissions in northeastern Indonesian Papua. Am J Trop Med Hyg. 2007, 77: 984-991.
Genton B, D'Acremont V, Rare L, Baea K, Reeder JC, Alpers MP, Muller I: Plasmodium vivax and mixed infections are associated with severe malaria in children: a prospective cohort study from Papua New Guinea. PLoS Med. 2008, 5: e127-10.1371/journal.pmed.0050127.
Kochar DK, Das A, Kochar SK, Saxena V, Sirohi P, Garg S, Kochar A, Khatri MP, Gupta V: Severe Plasmodium vivax malaria: a report on serial cases from Bikaner in northwestern India. Am J Trop Med Hyg. 2009, 80: 194-198.
Parakh A, Agarwal N, Aggarwal A, Aneja A: Plasmodium vivax malaria in children: uncommon manifestations. Ann Trop Paediatr. 2009, 29: 253-256. 10.1179/027249309X12547917868844.
Price RN, Tjitra E, Guerra CA, Yeung S, White NJ, Anstey NM: Vivax malaria: neglected and not benign. Am J Trop Med Hyg. 2007, 77 (6 Suppl): 79-87.
Tjitra E, Anstey NM, Sugiarto P, Warikar N, Kenangalem E, Karyana M, Lampah DA, Price RN: Multidrug-resistant Plasmodium vivax associated with severe and fatal malaria: a prospective study in Papua, Indonesia. PLoS Med. 2008, 5: e128-10.1371/journal.pmed.0050128.
Guerra CA, Howes RE, Patil AP, Gething PW, Van Boeckel TP, Temperley WH, Kabaria CW, Tatem AJ, Manh BH, Elyazar IR, Baird JK, Snow RW, Hay SI: The international limits and population at risk of Plasmodium vivax transmission in 2009. PLoS Negl Trop Dis. 2010, 4: e774-10.1371/journal.pntd.0000774.
Guerra CA, Snow RW, Hay SI: Defining the global spatial limits of malaria transmission in 2005. Adv Parasitol. 2006, 62: 157-179.
Guerra CA, Snow RW, Hay SI: Mapping the global extent of malaria in 2005. Trends Parasitol. 2006, 22: 353-358. 10.1016/j.pt.2006.06.006.
Baird JK: Resistance to therapies for infection by Plasmodium vivax. Clin Microbiol Rev. 2009, 22: 508-534. 10.1128/CMR.00008-09.
Sattabongkot J, Tsuboi T, Zollner GE, Sirichaisinthop J, Cui L: Plasmodium vivax transmission: chances for control?. Trends Parasitol. 2004, 20: 192-198. 10.1016/j.pt.2004.02.001.
Sautet J, Ranque J, Vuillet F, Vuillet J: Quelques notes parasitologiques sur le paludisme et l'anophélisme en Mauritanie. Med Trop (Mars). 1948, 8: 32-39.
Cortes H, Morillas-Marquez F, Valero A: Malaria in Mauritania: the first cases of malaria endemic to Nouakchott. Trop Med Int Health. 2003, 8: 297-300. 10.1046/j.1365-3156.2003.01029.x.
Gautret P, Legros F, Koulmann P, Rodier MH, Jacquemin JL: Imported Plasmodium vivax malaria in France: geographical origin and report of an atypical case acquired in Central or Western Africa. Acta Trop. 2001, 78: 177-181. 10.1016/S0001-706X(00)00181-9.
Lekweiry KM, Abdallahi MO, Ba H, Arnathau C, Durand P, Trape JF, Salem AO: Preliminary study of malaria incidence in Nouakchott, Mauritania. Malar J. 2009, 8: 92-10.1186/1475-2875-8-92.
Lekweiry KM, Basco LK, Salem MS, Hafid JE, Marin-Jauffre A, Weddih AO, Briolant S, Bogreau H, Pradines B, Rogier C, Trape JF, Boukhary AO: Malaria prevalence and morbidity among children reporting at health facilities in Nouakchott, Mauritania. Trans R Soc Trop Med Hyg. 2011,
Gelpi AP, King MC: Duffy blood group and malaria. Science. 1976, 191: 1284-10.1126/science.1257752.
Mercereau-Puijalon O, Menard D: Plasmodium vivax and the Duffy antigen: a paradigm revisited. Transfus Clin Biol. 2010, 17: 176-183. 10.1016/j.tracli.2010.06.005.
Miller LH, Mason SJ, Clyde DF, McGinniss MH: The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976, 295: 302-304. 10.1056/NEJM197608052950602.
Hadley TJ, Lu ZH, Wasniowska K, Martin AW, Peiper SC, Hesselgesser J, Horuk R: Postcapillary venule endothelial cells in kidney express a multispecific chemokine receptor that is structurally and functionally identical to the erythroid isoform, which is the Duffy blood group antigen. J Clin Invest. 1994, 94: 985-991. 10.1172/JCI117465.
Horuk R, Peiper SC: Chemokines: molecular double agents. Curr Biol. 1996, 6: 1581-1582. 10.1016/S0960-9822(02)70777-X.
Peiper SC, Wang ZX, Neote K, Martin AW, Showell HJ, Conklyn MJ, Ogborne K, Hadley TJ, Lu ZH, Hesselgesser J, Horuk R: The Duffy antigen/receptor for chemokines (DARC) is expressed in endothelial cells of Duffy negative individuals who lack the erythrocyte receptor. J Exp Med. 1995, 181: 1311-1317. 10.1084/jem.181.4.1311.
He W, Neil S, Kulkarni H, Wright E, Agan BK, Marconi VC, Dolan MJ, Weiss RA, Ahuja SK: Duffy antigen receptor for chemokines mediates trans-infection of HIV-1 from red blood cells to target cells and affects HIV-AIDS susceptibility. Cell Host Microbe. 2008, 4: 52-62. 10.1016/j.chom.2008.06.002.
Horne KC, Li X, Jacobson LP, Palella F, Jamieson BD, Margolick JB, Martinson J, Turkozu V, Visvanathan K, Woolley IJ: Duffy antigen polymorphisms do not alter progression of HIV in African Americans in the MACS cohort. Cell Host Microbe. 2009, 5: 415-417. 10.1016/j.chom.2009.04.013. author reply 418-419
Smolarek D, Hattab C, Hassanzadeh-Ghassabeh G, Cochet S, Gutierrez C, de Brevern AG, Udomsangpetch R, Picot J, Grodecka M, Wasniowska K, Muyldermans S, Colin Y, Le Van Kim C, Czerwinski M, Bertrand O: A recombinant dromedary antibody fragment (VHH or nanobody) directed against human Duffy antigen receptor for chemokines. Cell Mol Life Sci. 2010, 67: 3371-3387. 10.1007/s00018-010-0387-6.
Chaudhuri A, Polyakova J, Zbrzezna V, Williams K, Gulati S, Pogo AO: Cloning of glycoprotein D cDNA, which encodes the major subunit of the Duffy blood group system and the receptor for the Plasmodium vivax malaria parasite. Proc Natl Acad Sci USA. 1993, 90: 10793-10797. 10.1073/pnas.90.22.10793.
Collins WE, Nguyen-Dinh P, Sullivan JS, Morris CL, Galland GG, Richardson BB, Nesby S: Adaptation of a strain of Plasmodium vivax from Mauritania to New World monkeys and anopheline mosquitoes. J Parasitol. 1998, 84: 619-621. 10.2307/3284734.
Donahue RP, Bias WB, Renwick JH, McKusick VA: Probable assignment of the Duffy blood group locus to chromosome 1 in man. Proc Natl Acad Sci USA. 1968, 61: 949-955. 10.1073/pnas.61.3.949.
Horuk R, Chitnis CE, Darbonne WC, Colby TJ, Rybicki A, Hadley TJ, Miller LH: A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science. 1993, 261: 1182-1184. 10.1126/science.7689250.
Neote K, Mak JY, Kolakowski LF, Schall TJ: Functional and biochemical analysis of the cloned Duffy antigen: identity with the red blood cell chemokine receptor. Blood. 1994, 84: 44-52.
Rot A, Horuk R: The duffy antigen receptor for chemokines. Methods Enzymol. 2009, 461: 191-206.
Langhi DM, Bordin JO: Duffy blood group and malaria. Hematology. 2006, 11: 389-398. 10.1080/10245330500469841.
Tournamille C, Le Van Kim C, Gane P, Cartron JP, Colin Y: Molecular basis and PCR-DNA typing of the Fya/fyb blood group polymorphism. Hum Genet. 1995, 95: 407-410.
Mallinson G, Soo KS, Schall TJ, Pisacka M, Anstee DJ: Mutations in the erythrocyte chemokine receptor (Duffy) gene: the molecular basis of the Fya/Fyb antigens and identification of a deletion in the Duffy gene of an apparently healthy individual with the Fy(a-b-) phenotype. Br J Haematol. 1995, 90: 823-829. 10.1111/j.1365-2141.1995.tb05202.x.
Howes RE, Patil AP, Piel FB, Nyangiri OA, Kabaria CW, Gething PW, Zimmerman PA, Barnadas C, Beall CM, Gebremedhin A, Ménard D, Williams TN, Weatherall DJ, Hay SI: The global distribution of the Duffy blood group. Nat Commun. 2011, 2: 266-
Iwamoto S, Li J, Sugimoto N, Okuda H, Kajii E: Characterization of the Duffy gene promoter: evidence for tissue-specific abolishment of expression in Fy(a-b-) of black individuals. Biochem Biophys Res Commun. 1996, 222: 852-859. 10.1006/bbrc.1996.0833.
Iwamoto S, Omi T, Kajii E, Ikemoto S: Genomic organization of the glycoprotein D gene: Duffy blood group Fya/Fyb alloantigen system is associated with a polymorphism at the 44-amino acid residue. Blood. 1995, 85: 622-626.
Tournamille C, Colin Y, Cartron JP, Le Van Kim C: Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995, 10: 224-228. 10.1038/ng0695-224.
Zimmerman PA, Woolley I, Masinde GL, Miller SM, McNamara DT, Hazlett F, Mgone CS, Alpers MP, Genton B, Boatin BA, Kazura JW: Emergence of FY*A(null) in a Plasmodium vivax-endemic region of Papua New Guinea. Proc Natl Acad Sci USA. 1999, 96: 13973-13977. 10.1073/pnas.96.24.13973.
Albuquerque SR, Cavalcante Fde O, Sanguino EC, Tezza L, Chacon F, Castilho L, dos Santos MC: FY polymorphisms and vivax malaria in inhabitants of Amazonas State, Brazil. Parasitol Res. 2010, 106: 1049-1053. 10.1007/s00436-010-1745-x.
Kasehagen LJ, Mueller I, Kiniboro B, Bockarie MJ, Reeder JC, Kazura JW, Kastens W, McNamara DT, King CH, Whalen CC, Zimmerman PA: Reduced Plasmodium vivax erythrocyte infection in PNG Duffy-negative heterozygotes. PLoS One. 2007, 2: e336-10.1371/journal.pone.0000336.
Sellami MH, Kaabi H, Midouni B, Dridi A, Mojaat N, Boukef MK, Hmida S: Duffy blood group system genotyping in an urban Tunisian population. Ann Hum Biol. 2008, 35: 406-415. 10.1080/03014460802082127.
Chown B, Lewis M, Kaita H: The Duffy Blood Group System in Caucasians: Evidence for a New Allele. Am J Hum Genet. 1965, 17: 384-389.
Daniels GL, Anstee DJ, Cartron JP, Dahr W, Issitt PD, Jorgensen J, Kornstad L, Levene C, Lomas-Francis C, Lubenko A, Mallory D, Moulds JJ, Okubo Y, Overbeeke M, Reid ME, Rouger P, Seidl S, Sistonen P, Wendel S, Woodfield G, Zelinski T: Blood group terminology 1995. ISBT Working Party on terminology for red cell surface antigens. Vox Sang. 1995, 69: 265-279. 10.1111/j.1423-0410.1995.tb02611.x.
Olsson ML, Smythe JS, Hansson C, Poole J, Mallinson G, Jones J, Avent ND, Daniels G: The Fy(x) phenotype is associated with a missense mutation in the Fy(b) allele predicting Arg89Cys in the Duffy glycoprotein. Br J Haematol. 1998, 103: 1184-1191. 10.1046/j.1365-2141.1998.01083.x.
Pogo AO, Chaudhuri A: The Duffy protein: a malarial and chemokine receptor. Semin Hematol. 2000, 37: 122-129. 10.1016/S0037-1963(00)90037-4.
Tournamille C, Le Van Kim C, Gane P, Le Pennec PY, Roubinet F, Babinet J, Cartron JP, Colin Y: Arg89Cys substitution results in very low membrane expression of the Duffy antigen/receptor for chemokines in Fy(x) individuals. Blood. 1998, 92: 2147-2156.
Yazdanbakhsh K, Rios M, Storry JR, Kosower N, Parasol N, Chaudhuri A, Reid ME: Molecular mechanisms that lead to reduced expression of duffy antigens. Transfusion. 2000, 40: 310-320. 10.1046/j.1537-2995.2000.40030310.x.
Gassner C, Kraus RL, Dovc T, Kilga-Nogler S, Utz I, Mueller TH, Schunter F, Schoenitzer D: Fyx is associated with two missense point mutations in its gene and can be detected by PCR-SSP. Immunohematology. 2000, 16: 61-67.
Parasol N, Reid M, Rios M, Castilho L, Harari I, Kosower NS: A novel mutation in the coding sequence of the FY*B allele of the Duffy chemokine receptor gene is associated with an altered erythrocyte phenotype. Blood. 1998, 92: 2237-2243.
Reid ME, Rios M, Roye K, Chaudhuri A, Pogo O, Yazdanbakhsh K, Coghlan G, Kosower N, Parasol N: Molecular basis of FYX. Transfusion. 1998, 38 (102S):
Castilho L, Rios M, Pellegrino J, Saad ST, Costa FF, Reid ME: A novel FY allele in Brazilians. Vox Sang. 2004, 87: 190-195. 10.1111/j.1423-0410.2004.00554.x.
Cavasini CE, de Mattos LC, Couto AA, Couto VS, Gollino Y, Moretti LJ, Bonini-Domingos CR, Rossit AR, Castilho L, Machado RL: Duffy blood group gene polymorphisms among malaria vivax patients in four areas of the Brazilian Amazon region. Malar J. 2007, 6: 167-10.1186/1475-2875-6-167.
Cavasini CE, Mattos LC, Couto AA, Bonini-Domingos CR, Valencia SH, Neiras WC, Alves RT, Rossit AR, Castilho L, Machado RL: Plasmodium vivax infection among Duffy antigen-negative individuals from the Brazilian Amazon region: an exception?. Trans R Soc Trop Med Hyg. 2007, 101: 1042-1044. 10.1016/j.trstmh.2007.04.011.
Menard D, Barnadas C, Bouchier C, Henry-Halldin C, Gray LR, Ratsimbasoa A, Thonier V, Carod JF, Domarle O, Colin Y, Bertrand O, Picot J, King CL, Grimberg BT, Mercereau-Puijalon O, Zimmerman PA: Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci USA. 2010, 107: 5967-5971. 10.1073/pnas.0912496107.
Mendes C, Dias F, Figueiredo J, Mora VG, Cano J, de Sousa B, do Rosario VE, Benito A, Berzosa P, Arez AP: Duffy Negative Antigen Is No Longer a Barrier to Plasmodium vivax - Molecular Evidences from the African West Coast (Angola and Equatorial Guinea). PLoS Negl Trop Dis. 2011, 5: e1192-10.1371/journal.pntd.0001192.
Ryan JR, Stoute JA, Amon J, Dunton RF, Mtalib R, Koros J, Owour B, Luckhart S, Wirtz RA, Barnwell JW, Rosenberg R: Evidence for transmission of Plasmodium vivax among a duffy antigen negative population in Western Kenya. Am J Trop Med Hyg. 2006, 75: 575-581.
NCBI/Primer-Blast. [http://www.ncbi.nlm.nih.gov/tools/primer-blast/]
Galinski MR, Barnwell JW: Plasmodium vivax: who cares?. Malar J. 2008, 7 (Suppl 1): S9-10.1186/1475-2875-7-S1-S9.
Mueller I, Galinski MR, Baird JK, Carlton JM, Kochar DK, Alonso PL, del Portillo HA: Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis. 2009, 9: 555-566. 10.1016/S1473-3099(09)70177-X.
Lekweiry KM, Boukhary AOMS, Gaillard T, Wurtz N, Bogreau H, Hafid JE, Trape JF, Bouchiba H, Salem MSOA, Pradines B: Molecular surveillance of drug-resistant Plasmodium vivax using pvdhfr, pvdhps and pvmdr1 markers in Nouakchott, Mauritania. J Antimicrob Chemother. 2011,
Lepers JP, Simonneau M, Charmot G: [The Duffy blood group system in the population of Nouakchott (Mauritania)](in French). Bull Soc Pathol Exot Filiales. 1986, 79: 417-420.
Jeddi Blouza A, Loukil I, Mhenni A, Ben Rayana C, Hmida S: [Blood groups and open-angle glaucoma in Tunisia](in French). J Fr Ophtalmol. 2007, 30: 493-496. 10.1016/S0181-5512(07)89629-X.
Fernandez-Santander A, Kandil M, Luna F, Esteban E, Gimenez F, Zaoui D, Moral P: Genetic relationships between southeastern Spain and Morocco: New data on ABO, RH, MNSs, and DUFFY polymorphisms. Am J Hum Biol. 1999, 11: 745-752. 10.1002/(SICI)1520-6300(199911/12)11:6<745::AID-AJHB4>3.0.CO;2-W.
Acknowledgements and funding
The authors thank the direction and the staff of the National Hospital, the Chiekh Zayed Hospital and the District Health Center of Teyarett for their aid in recruiting patients and the patients for kindly agreeing to participate in the study.
The authors are grateful to the programme "For Women in Science" granted by L'Oréal-UNESCO foundation and the French Centre for the welcome and the international trade.
This study was supported by the Délégation Générale pour l'Armement and the Direction Centrale du Service de Santé des Armées (grant no. 10co404).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SB, NW, LKB, JEH, MSOAS, BP, JFT and CR conceived and designed the experiments. KML and NW performed the genotyping of Duffy gene and the diagnosis of Plasmodium vivax. HB, AOMSB, SB and KML contributed to reagents/materials/analysis tools. SB, NW and HB analysed the data. NW, SB, LKP and BP wrote the paper. All authors read and approved the final manuscript.
Electronic supplementary material
12936_2011_1922_MOESM1_ESM.DOC
Additional file 1: Duffy blood group nomenclature. Duffy alleles and their corresponding genotype and phenotypic and expression. (DOC 28 KB)
12936_2011_1922_MOESM2_ESM.DOC
Additional file 2: Phenotype expression relative to 32 different genotypes possible from eight known Duffy alleles (FYA, FYB, FYB*, FYX1, FYX2, FYX3, FYAES and FYBES). (DOC 66 KB)
12936_2011_1922_MOESM3_ESM.XLS
Additional file 3: Individual data, Plasmodium diagnosis and Duffy blood group genotypes for the patients selected in the study. (XLS 92 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Wurtz, N., Mint Lekweiry, K., Bogreau, H. et al. Vivax malaria in Mauritania includes infection of a Duffy-negative individual. Malar J 10, 336 (2011). https://doi.org/10.1186/1475-2875-10-336
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-10-336