Abstract
Background
Planting tested forest reproductive material is crucial to ensure the increased resilience of intensively managed productive stands for timber and wood product markets under climate change scenarios. Single-step Genomic Best Linear Unbiased Prediction (ssGBLUP) analysis is a cost-effective option for using genomic tools to enhance the accuracy of predicted breeding values and genetic parameter estimation in forest tree species. Here, we tested the efficiency of ssGBLUP in a tropical multipurpose tree species, Cordia africana, by partial population genotyping. A total of 8070 trees from three breeding seedling orchards (BSOs) were phenotyped for height. We genotyped 6.1% of the phenotyped individuals with 4373 single nucleotide polymorphisms. The results of ssGBLUP were compared with pedigree-based best linear unbiased prediction (ABLUP) and genomic best linear unbiased prediction (GBLUP), based on genetic parameters, theoretical accuracy of breeding values, selection candidate ranking, genetic gain, and predictive accuracy and prediction bias.
Results
Genotyping a subset of the study population provided insights into the level of relatedness in BSOs, allowing better genetic management. Due to the inbreeding detected within the genotyped provenances, we estimated genetic parameters both with and without accounting for inbreeding. The ssGBLUP model showed improved performance in terms of additive genetic variance and theoretical breeding value accuracy. Similarly, ssGBLUP showed improved predictive accuracy and lower bias than the pedigree-based relationship matrix (ABLUP).
Conclusions
This study of C. africana, a species in decline due to deforestation and selective logging, revealed inbreeding depression. The provenance exhibiting the highest level of inbreeding had the poorest overall performance. The use of different relationship matrices and accounting for inbreeding did not substantially affect the ranking of candidate individuals. This is the first study of this approach in a tropical multipurpose tree species, and the analysed BSOs represent the primary effort to breed C. africana.
Similar content being viewed by others
Background
In the tropical world, there is a need to undertake forest and landscape restoration to improve degraded former forest lands [1]. Combined with climate change, this poses huge challenges to both the amount and quality of the plant material used for this restoration. Consequently, there is a need for breeding and forest tree improvement of a multitude of species. Globally, large-scale reforestation and afforestation programmes have focused on a few genera and species, mainly conifers, such as Pinus spp. and Picea spp., or broadleaves, such as Eucalyptus spp. and Acacia spp., often involving intercontinental movement and monoculture plantations [2]. However, because of the risk of genetic diversity loss and the capacity to adapt to extreme climates, as well as to other ecological and socioeconomic issues [3], the use of multiple/diverse species seems to be a more realistic approach [4]. Conventional breeding is resource-intensive for managing and tracking the seed-generated families in the nursery and the progeny tests. Furthermore, in a conventional first-generation breeding programme, early selection is inevitably based on juvenile traits. This emphasises the importance of testing the possibility of using genomics in pedigree reconstruction and the estimation of additive relationships in natural populations of tropical tree species. This study considers the breeding of multipurpose tropical species of local/regional importance using genomic evaluations via a genotyping-by-sequencing approach, that is, DArT partial genome sequencing, which does not rely on the availability of genomic resources for the species.
Experimental plant genetics and breeding programmes rely on the ability to predict and visualise the inheritance of alleles underlying traits of interest [5]. To estimate genetic parameters (i.e., variance components and heritability), it is necessary to infer additive genetic relationships among individuals based on their known pedigrees [6]. Classical quantitative genetics uses this information on additive relationships to estimate heritability, covariance between traits, and genotype-by-environment interactions. In addition to enabling the estimation of genetic parameters, pedigree information is also critical for maintaining high genetic variation and low levels of inbreeding. These latter issues are critical for populations to face environmental changes and ensure long-term genetic gain [7]. However, there is a limitation related to the accuracy and completeness of the available pedigree information, especially in wild populations.
In the absence of pedigree information, molecular marker data have been utilised to reconstruct pedigrees since the 1970s and 1980s [8,9,10,11]. In forest trees, pedigree reconstruction via DNA markers and subsequent quantitative genetic analyses as a breeding approach was introduced in the 2000’s [12,13,14,15], but these first studies were based on using only a few highly variable microsatellite DNA markers. Despite the obvious potential for tree breeding via pedigree reconstruction, there are issues that need to be considered. One such issue is the incompleteness of sampling potential parents, as pedigree and/or sibship reconstruction based on a few DNA markers requires information on the parental population (see [16] for instance). This is particularly a challenge when one works with natural populations or with plantations established with commercial seed lots. In addition, even with successful pedigree or sib-ship reconstruction, there is a problem of hidden relatedness because the focus is only on one-generation relatedness, where each family is considered unrelated and no Mendelian sampling variance is considered. The accuracy of genetic parameter estimation and rankings of predicted breeding values can be affected by hidden relatedness [17,18,19,20].
Accurate estimation of relatedness between individuals in breeding populations using DNA markers is important not only to precisely estimate genetic parameters but also for effective inbreeding management [21]. Furthermore, DNA markers can assess the genetic diversity of the entire population across different gene pools [22].
Cordia africana is a fast-growing tree species that is highly valued in Ethiopia for its timber. As one of the most commercially utilised species, it plays an important role in generating household income from the sale of wood products [23]. In its current distribution, the populations of C. africana are heavily affected by deforestation, fragmentation, and selective logging. The northern part of Ethiopia has been extremely deforested. As a result, this species is mainly represented by scattered trees on farmlands, church compounds, and graveyards, while a relatively continuous forest only exists in a few spots [24, 25]. Cordia africana is an indigenous tree species that has been identified and given conservation priority nationwide in Ethiopia [26]. Moreover, C. africana is a priority species included in the tree breeding programme under the Provision of Adequate Tree Seed Portfolio (PATSPO), supporting afforestation efforts in Ethiopia [27]. This breeding programme of the C. africana is based on the establishment of breeding seedling orchards (BSOs). The improved seed produced by the BSOs is foreseen to play a significant role in species conservation, promoting its sustainable use by enabling the establishment of improved C.africana plantations for different end uses and thereby decreasing the pressure on natural populations. As the first generation of breeding this species, seeds were collected from the major growing areas of the country, with the aim of broadening the genetic basis and ensuring continuous gain.
The mating system of C. africana has not yet been specifically documented. However, as a long-living tree with an efficient means of pollen and seed dispersal, the species is speculated to be predominantly outcrossing [25]. Nonetheless, due to its hermaphrodite flowers [23], and no evidence of self-incompatibility, self-fertilisation might be possible under some conditions. Moreover, even in species that are not self-compatible, inbreeding can occur through mating between related individuals. As a species with a fragmented/scattered distribution and exposure to selective/illegal logging, the probability of building up co-ancestry is high. This makes it difficult to obtain a reliable estimate of genetic parameters using the conventional pedigree-based approach and increases the risk of selecting related materials. In recent years, the utilisation of realised genetic relationships has been shown to produce a more accurate estimation of genetic parameters by capturing within-family variation that arises from Mendelian segregation (e.g., [28]). This is because the method enables the detection of the realised genetic covariance based on a fraction of the genome that is identical by descent or by state between individuals [29]. However, since C. africana is not a model species and lacks genetic information, and because there are few resources allocated to the breeding programme, the expense of using large-scale genotyping in the operational breeding of this species is still not feasible. To overcome this, single-step genomic best linear unbiased prediction (ssGBLUP), an approach in which the realised genomic relatedness of a small portion of genotyped individuals is combined with a large proportion of non-genotyped individuals in a single genetic evaluation, has been proposed as an effective analytical method [30,31,32]. The ssGBLUP method combines the pedigree-based A-matrix of non-genotyped individuals with the G-matrix of genotyped individuals into a single genetic covariance hybrid H-matrix. This method has been demonstrated to be an efficient option for improving the derived genetic parameters’ precision and breeding value accuracy from the actual generation (e.g., [33,34,35]) and for genomic prediction (e.g., [36,37,38,39]) in different tree species.
The aim of this study was to evaluate the efficiency of ssGBLUP, with minimum genotyping effort, in C. africana breeding. This study compared the results of ssGBLUP with pedigree-based best linear unbiased prediction (ABLUP) and genomic best linear unbiased prediction (GBLUP), based on various factors, including genetic parameters, theoretical accuracy of breeding values, selection candidate ranking, genetic gain, efficiency of the ssGBLUP method, and predictive accuracy and prediction bias. Additionally, by genotyping a subset of the study population, we aimed to determine the level of relatedness in the three BSOs and the potential effects of inbreeding on C. africana. This knowledge will be useful in better managing the genetic resources of C. africana. While previously mentioned studies have demonstrated the utility of ssGBLUP in tree breeding, this is the first report of the use of genomic tools in C. africana breeding. Finally, this is the first study of this approach in a tropical multipurpose tree species, and the analysed BSOs represent the primary effort to breed C. africana.
Results
Population and family structure
The pairwise relationship coefficients of the G-matrix showed a clear provenance and family structure (Fig. 1a). The heatmap revealed separate grouping of the genotyped provenances, which coincided with the geographic distance between their origins (Supplementary Table S1). Provenance P30 included 161 trees from the North Bench zone (southern Ethiopia), provenance P31 included 212 trees from Adwa (northern Ethiopia), and provenance P34 included 121 trees from Harar (eastern Ethiopia). Similarly, principal component analysis of genotypes showed grouping of individuals according to these three provenances (Fig. 1b).
Heatmap of pairwise genomic relationship coefficients among the 490 genotyped C. africana trees (a), principal component analysis of the three genotyped provenances (b) and density plot of the four additive realised relationship coefficients within the G-matrix (c). In plot (a), the heat scale represents the degree of pairwise relationship coefficients for all pairs of trees. It is based on the provided pedigree (A-matrix before pedigree correction) in the upper diagonal and the realized genomic relationship coefficients (G-matrix) in the lower diagonal. The three light blue triangles are placed diagonally along the lower diagonal, indicating moderate relatedness between individuals within a provenance in the serial order of P30 (the first triangle), P31 (the second triangle), and P34 (the third triangle) from left to right. The smaller green boxes along the diagonal represent the families within the provenances based on the A- and G-matrix, on the upper and lower sides of the diagonal, respectively. The G-matrix (lower diagonal) also shows pedigree errors in 15 individuals that appear to belong to different families, either within the same or different provenances. In plot (b), the first principal component (PC1) is plotted against the second principal component (PC2), and different colours represent different provenances: P30 in red, P31 in green, and P34 in blue. Plot (c) displays four peaks representing different levels of realised genetic relationships between individuals. The first peak (highest) shows the relationship between individuals from different provenances. The second peak represents the relationship between unrelated individuals among provenances and within provenances. The third peak represents the relationship between individuals from different families within the same provenance, and the fourth peak attached to the third peak represents the relationship within families
By examining the genomic pairwise relatedness among the 490 trees, we found various levels of genetic relationships within the population. The estimated relatedness between individuals, obtained by genetic marker analysis, did not always match the expected values, such as half siblings (expected value 0.25), and unrelated individuals (expected value 0.00). However, there was only little variation in these estimates across the different provenances. For the 490 trees, the average relatedness for unrelated individuals within provenance was above zero for most pairs and close to an expected half-sib relationship of 0.25 for many pairs that are assumed to be unrelated. The average minimum pairwise genomic relatedness between trees belonging to the same open-pollinated family was 0.28, while the average across all families in all three provenances was 0.39. In contrast, the genomic relatedness value between individuals from different provenances was negative for most pairs (Fig. 1c).
Inbreeding and provenance performance
According to the diagonal elements of the G-matrix, two of the three provenances showed an inbreeding coefficient Fi above zero. Fi was calculated by subtracting one from the diagonal elements of the G-matrix [40]. Genotyped provenance P31 had the highest Fi value (0.54), while provenance P34 had the lowest (0.01). Provenance P30 had a Fi of 0.29. The average Fi across all genotyped provenances was 0.33. The best linear unbiased estimates (BLUE) from the ssGBLUP model of the genotyped provenances revealed that their height was consistent with their respective levels of inbreeding. Accordingly, P34 was highest, while P31 was lowest across all sites (Table 1).
To further examine the inbreeding effect, we conducted a correlation test between mean family height and mean family inbreeding coefficient. Overall, there was a strong indication of a negative effect of inbreeding on height (r = − 0.82; Fig. 2).
Additive genetic variances and heritability estimates
Without accounting for inbreeding, the additive variance estimates (\({\sigma }_{a}^{2}\)) for height using the A-matrix (ABLUP-1) ranged from 2.32 in the SM site to 3.21 in the Suba site, while the estimates from the ssGBLUP-1 ranged from 2.63 in the ILRI site to 3.75 at Suba. When accounting for inbreeding, the additive variance using the A-matrix (ABLUP-2) was reduced to 1.73 in ILRI, 1.52 in the SM site, and 2.24 in the Suba site (Table 2). The estimates from ssGBLUP-2 were 2.05 in ILRI, 2.04 in SM, and 2.93 in Suba. Although incorporating a selfing rate resulted in a considerable decrease in additive genetic variance and heritability in both the ABLUP and ssGBLUP models, the discrepancy was larger between the two ABLUP models than between the two ssGBLUP models. At the ILRI site, accounting for inbreeding decreased the additive genetic variance by 36.4% in ABLUP and 22.1% in ssGBLUP. In SM, accounting for inbreeding decreased the additive variance estimates by 34.5% and 23.9% in ABLUP and ssGBLUP, respectively. The Suba site showed the same pattern, with the estimate in ABLUP decreasing by 33.6% and the estimate in ssGBLUP decreasing by 21.9%. Generally, there was a constant increase in additive variance estimates when moving from ABLUP to ssGBLUP at all sites, except for the without-inbreeding scenario in the ILRI site, where it slightly decreased from 2.72 in ABLUP to 2.63 in ssGBLUP.
As expected, the narrow-sense heritability estimates of the ABLUP and ssGBLUP models mirrored those of the additive genetic variance estimates in both scenarios at all sites. The highest estimates were observed for the Suba site in all models studied compared to the same models at the other sites. Generally, ssGBLUP showed an improved heritability compared to its counterpart ABLUP within the same scenario, except for the scenario without inbreeding at the ILRI site, where ABLUP showed a slightly higher estimate (0.26) compared to ssGBLUP (0.25) (Table 2).
Additive genetic correlations across sites
Additive genetic correlations between sites from all multiple-site models varied from 0.40 to 0.53 (see Supplementary Fig. S1 for details). ILRI and SM showed a slightly higher correlation in all models. The lowest average across-model genetic correlation between sites (0.44) was observed between Suba and ILRI. The moderate genetic correlations between sites indicates the presence of a genotype-by-environment interaction.
Theoretical accuracy of breeding values
Overall, the highest average theoretical accuracy of the predicted breeding values was observed for ssGBLUP-1 in Suba (0.68 ± 0.02), followed by GBLUP for genotyped individuals in ILRI (0.65 ± 0.04) (Table 3). The use of genomic relatedness did not only improve the accuracy of breeding values for trees from genotyped families but also for trees from families with no genotyped individuals. In the inbreeding scenario, the genotyped site IRLI showed that ssGBLUP-2 improved the breeding value accuracy by 12% compared to ABLUP-2. This increment was higher for SM (17%) and Suba (16.4%) sites. The “without inbreeding” scenario showed the same pattern, except for the ILRI site, where theoretical accuracy decreased marginally by 1.7%. As expected, given the higher estimates of the additive genetic variances (Table 2), the ABLUP and ssGBLUP models without inbreeding showed higher theoretical accuracies than the respective models “with inbreeding”.
Efficiency
Efficiency (\(E\)) in the accuracy of predicted breeding values, i.e., the proportion of extra benefit obtained from using ssGBLUP calculated for the genotyped site (ILRI) [41], was 0.4 for the inbreeding scenarios. Thus, assuming that GBLUP was 100% more efficient than ABLUP, ssGBLUP was 40% more efficient than ABLUP when inbreeding was considered. However, in the scenario without inbreeding, due to the lower theoretical accuracy of the breeding values (because of lower additive genetic variance) of ssGBLUP at the ILRI site, ssGBLUP had no advantage over ABLUP.
Candidate ranking and expected genetic gain
The proportion of common selection candidates in the top 10% of trees was used to test the impact of including genomic information and inbreeding in the prediction of breeding values. The lowest proportion of common candidates (96.2%) was observed between the ABLUP-1 (without inbreeding) and ssGBLUP-2 (with inbreeding) models. The two ssGBLUP models showed 96.6% common candidates. The two ABLUP models showed a similar proportion of shared candidates (96.5%). The ABLUP and ssGBLUP models without considering inbreeding (i.e., ABLUP-1 vs. ssGBLUP-1) showed a large proportion of common candidates (98.2%), followed by the same models in the inbreeding scenario (97.9%). Thus, neither the change in the model nor accounting for inbreeding had a substantial effect on the ranking of the top 10% of candidates. However, there were still some changes in the ranking of individuals. Supplementary Fig. S2 shows the change in ranks for the top 50 individuals between the models.
The proportion of selected candidates from different provenances in the top 10% was used to determine the impact of inbreeding (Table 4). The provenance with the highest inbreeding (P31, Fi = 0.54) had the lowest proportion of selection candidates in the top 10% (T10%, Table 4), while the provenance with the least inbreeding (P34, Fi = 0.01) had the highest proportion of selection candidates. To further check for signs of inbreeding depression, we also examined the proportion of the genotyped provenances among the worst-performing individuals (in the lowest 10% of breeding values) (Table 4). In all models, we found that over 98% of the low-ranked individuals came from the provenance with the highest level of inbreeding (P31).
Finally, the expected genetic gains from the top-ranked 10% were 57.5% and 57.6% for ABLUP-1 and ssGBLUP-1, respectively. The models in the inbreeding scenario showed slightly lower genetic gain, i.e., 55.7% for ABLUP-2 and 56.8% for ssGBLUP-2, over the original population mean.
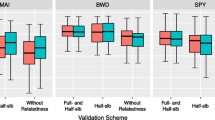
Predictive accuracy and prediction bias of the models
Overall, the ssGBLUP prediction models showed the highest predictive accuracy (PA) compared to the ABLUP and GBLUP models for all studied scenarios (i.e., with and without inbreeding, and random and within provenance cross-validation). From the two cross-validation scenarios (i.e., random and within provenance), the PA obtained from within provenance cross-validation was higher than the PA obtained from random cross-validation (Fig. 3). Overall, the inbreeding scenario showed a higher PA than non-inbreeding. In this sense, the ssGBLUP model in the inbreeding scenario (ssGBLUP-2) combined with within-provenance sampling showed the highest predictive accuracy (0.73 ± 0.02) (Supplementary Table S2), with almost similar outcome as the ABLUP model in the same scenario (ABLUP-2, 0.72 ± 0.02). Meanwhile, the lowest PA was observed for the GBLUP model in random cross-validation (0.53 ± 0.11). In all cases, both the ssGBLUP and ABLUP models showed a lower prediction bias (a value of ~ 1), while the GBLUP model had the highest bias in random cross-validation scenarios (0.79 ± 0.23). This might be attributed to the relatively small number of trees in the training and validation population (490 trees), which could have affected the model’s performance.
Average predictive accuracy (PA) and prediction bias (PB) for the ABLUP, GBLUP, and ssGBLUP prediction models studied using the four combined scenarios. These scenarios included the presence (I) or absence (NI) of inbreeding and random (R) and within-provenance (P) cross-validation scenarios. NOTE: I + P = Model with inbreeding combined with within provenance cross-validation; I + R = Model with inbreeding combined with random cross-validation; NI + P = Model without inbreeding combined with within provenance cross-validation; NI + R = Model without inbreeding combined with random cross-validation
Discussion
To ensure increased resilience of managed production plantations under climate change, it is crucial to plant tested forest reproductive material [42]. The availability of improved material serves a dual purpose; meeting the demand for timber and wood products but also relieving the pressure on the natural forests and helping restore locally endangered species. This is particularly true for C. africana in Ethiopia, where the habitats and populations are severely affected by deforestation, fragmentation, and selective logging. Thus, overcoming the deforestation issue while meeting the local demand for timber and wood products requires a supply of genetically diverse, healthy, and productive material. In this regard, the use of quantitative genetics methodology in forest tree breeding is widely recognised for its ability to deliver results [43]. However, there is limited time and resources to start traditional long-term breeding programmes to genetically improve the species. Incorporating genomic tools into practical tree breeding has been reported to increase gain by reducing the breeding cycle and improving selection efficiency [44].
This study tested the efficiency of breeding C. africana with the ssGBLUP approach using minimum genotyping effort and compared it to the ABLUP and GBLUP approaches. Comparisons included additive genetic variance, accuracy of the predicted breeding values, ranking of candidates for selection, genetic gain, efficiency of the ssGBLUP approach, and predictive accuracy and prediction bias. In addition, genotyping a subset of the study population provided insight into the degree of relatedness within the BSOs. This enabled evaluation of the potential impact of inbreeding in C. africana and enhanced our understanding of relatedness in the BSOs, which is useful for genetic management. This may imply possible restrictions on selection to avoid significant decrease in genetic variation. Restrictions that would otherwise not be implemented due to a lack of knowledge of relatedness in the material.
The pairwise genomic relationship coefficients of the C. africana genotyped trees differed from the expected pedigree-based values for half-sib and unrelated individuals. However, there was a clear differentiation among provenances, as individuals from different provenances appeared to be unrelated. The increased pairwise half-sib values (i.e., expected values 0.25 vs. realised values averaged across families 0.39) could be due to: 1) the build-up of co-ancestry and gene flow restrictions resulting from fragmented distribution of the species due to exposure to selective logging; and 2) background inbreeding due to lack of sufficient genetic separation, primarily due to close distance between the selected mother trees. This means that assuming an inbreeding level of zero, as is done in the classical breeding approach, could lead to an overestimation of genetic parameters, i.e., additive variance and heritability, and underestimation of relatedness across the studied sites. In this study, accounting for inbreeding decreased the additive variance estimate and heritability. Inflated heritability estimates without proper consideration of inbreeding concur with the results of other studies [45]. Moreover, hidden relatedness has been reported to result in upwardly biased estimates of heritability caused by the overestimation of additive genetic variance due to the unrealistic assumption of pure half-sibling relatedness within open-pollinated families, as well as the absence of historical relatedness among the parents [17, 46, 21].
In our study, the ssGBLUP models showed a constant increase in additive variance estimates compared to the ABLUP models at all sites, except for the “without inbreeding” scenario at the ILRI site. This is in line with results reported for growth traits in other tree species, for example, Eucalyptus [47, 48], lodgepole pine [38], white spruce [49], and loblolly pine [50]. However, studies on other tree species have reported that models using genomic evaluation (GBLUP or ssGBLUP) show decreased or similar additive variance estimates compared to models using pedigree-based information only for growth and wood quality traits [51,52,53]. The difference in genetic variance estimates between the pedigree and genomic-based relationship matrix could be because pedigree and genomic-based relationship matrices pertain to separate base populations, with genomic relationships reflecting the genotyped population and pedigree relationships reflecting the founders of the population under study [54].
The ssGBLUP models have an advantage over the ABLUP models in that they use the realised genomic relationship among individuals, whereas the ABLUP models are completely dependent on the assumed pedigree structure created by mating designs. The accuracy of the predicted breeding values is of great importance to tree breeders, and improvement in accuracy can be achieved by using genetic markers in the evaluation [33]. Here, the GBLUP showed the highest mean theoretical accuracy for breeding values. In all cases, the mean breeding value accuracy increased from ABLUP to ssGBLUP across the investigated genotyped and non-genotyped sites. Similar findings were reported in studies of forest trees (e.g., [34, 35, 37, 55]), mostly because of a smaller prediction error variance in ssGBLUP.
The genetic architecture of the trait, the choice of model, and the density of available markers in the model are factors that affect the accuracy of genomic prediction [56]. The use of genetic markers, capturing the additive relatedness among individuals, also captures the linkage disequilibrium (LD) between the SNPs and quantitative trait loci (QTLs), which affects the accuracy of the genomic estimated breeding values [57, 58]. The ssGBLUP provides a useful framework for combining the DNA marker data of the genotyped portion of the test population with the A-matrix used in BLUP analyses. As a result, it serves as a compromise between ABLUP and GBLUP [38]. In both tested cross-validation scenarios, i.e., random and within provenance cross-validation, the ABLUP and ssGBLUP models showed high PA and low PB, while GBLUP showed the lowest PA and highest PB. The reason for the lowest PA and higher PB in GBLUP could be because only 6.1 percent of the entire study population was genotyped. Consequently, we had substantially smaller training and validation sets. As previously observed in other forest trees using simulation [59] and empirical datasets [47, 60], the PA improved with the increasing size of the training set [39]. The within-provenance cross-validation scenarios showed a slightly higher average PA for the three predictive models studied. The three provenances in this study appeared to be unrelated. Moreover, our results showed that models in which inbreeding was considered showed improved PA. Increased relatedness between the training and validation populations is known to increase the model’s PA [61]. Thus, improved PA in the case of both within-provenance cross-validation and inbreeding scenarios could be attributed to the higher relatedness between the training and validation populations. Genetic diversity within the training population has also been reported to influence PA [62, 63].
Genetic diversity management is an integral part of tree breeding [64]. Retaining genetic diversity is crucial for long-term genetic gain, and also limiting inbreeding and hence inbreeding depression [65, 66]. Currently, C. africana populations in Ethiopia are severely affected by deforestation, fragmentation, and selective logging. Forest loss and fragmentation are known to change the landscape’s connectedness and composition [67]. A few examples of how these changes may impact genetic diversity include instances linked to decreased population sizes and the isolation of residual populations, which can affect and limit gene flow and cause the direct loss of genes [68]. The BSOs in this study were established by taking the necessary precautions to avoid inbreeding, i.e., by selecting mother trees with a minimum of 100 m distance from each other and in areas with a larger number of trees when possible. However, since the selfing rate might vary among species, populations, and individuals, it should be estimated using DNA marker data from the population under study [48]. This is especially true for C. africana in Ethiopia, where the fragmentation makes diversity management tricky. This is evident from the variation in the level of inbreeding between the genotyped provenances, with a mean inbreeding level of 0.33. Although it is difficult to conclude that C. africana suffers from inbreeding depression based on limited provenances, the most inbred provenance (P31, Fi = 0.54) showed signs of inbreeding depression, with the lowest overall performance. It also had the lowest proportion of selection candidates in the top 10%. In contrast, the least inbred provenance (P34, Fi = 0.01) showed the best overall performance. Similarly, the correlation between height and inbreeding at the family level (r = − 0.82, p < 0.0001) was a strong indicator of the negative influence of inbreeding. However, there are possible confounding effects of provenances and inbreeding, and the correlation between height and the inbreeding coefficient seems weaker and inconclusive within provenances. This could be due to the relatively small differences in the inbreeding coefficients within provenances.
A comparison of breeding value rankings from different models revealed change in ranks between the ABLUP and ssGBLUP models. Nevertheless, all models shared most of their top 10% of trees. The lowest proportion of common candidates in the top 10% of trees was between ABLUP-1 and ssGBLUP-2 (96.2%). This was expected because these two models represent scenarios without any genetic information and scenarios with all genetic information available. This maximises the information gap between the ABLUP-1 and ssGBLUP-2 models, resulting in a lower proportion of common selection candidates. The proportion of common candidates between the two ABLUP models with or without considering inbreeding was 96.5%, indicating that accounting for inbreeding has a minimal impact on the ranking of selection candidates. Generally, although the 10% selection scenarios using different approaches largely showed common candidate trees, this could, to some extent, be due to high inbreeding levels and depression. Differences in selected candidates between ABLUP-1 and ssGBLUP-2 would likely have been higher in the case of lower inbreeding without inbreeding depression. This means that the gap in the inbreeding level between provenances and the resulting impact on individual performance contributed to the selection of similar candidates across the models.
This study stresses the importance of sampling from multiple populations of native tree species, such as C. africana, having small, scattered populations with discontinuous tree distributions to enhance genetic diversity in BSOs. This is further emphasised because of the genotype-by-environment interactions indicated by moderate genetic correlations across the BSOs, despite being situated at almost similar altitudes. The differential performance of the species at the three BSOs could be attributed to the site differences in terms of soil nutrient, windiness, and other macro- and micro-climatic conditions. This species tends to struggle to grow in windy areas. Thus, the fact that Suba and SM sites are located on hilly sites could be the reason for the relatively slow early growth at these sites compared to ILRI.
Only 33.8% of the families had genotyped individuals. Genotyping individuals from all families could potentially have additional benefits for ssGBLUP models. Under fixed costs, this can be achieved by decreasing the number of individuals per family. We suggest conducting an in-depth population genetic analysis with more provenances to confirm whether the inbreeding level observed in the study is representative of C. africana populations in the country. Moreover, understanding the current genetic diversity of the species is crucial in devising appropriate measures for its conservation and sustainable utilisation.
Conclusion(s)
We tested the efficiency of ssGBLUP with minimum genotyping effort in genetic management and breeding of C. africana in BSOs. Following the inbreeding detected in two of the three genotyped provenances, we compared the ABLUP and ssGBLUP models with and without accounting for inbreeding. Although both evaluation models, with or without accounting for inbreeding, had similar top 10% selection candidates, the ssGBLUP model displayed better performance in terms of additive genetic variance, theoretical breeding value accuracy, predictive accuracy, and prediction bias. Therefore, the ssGBLUP model was more reliable and accurate in determining breeding values, and as a result, the likelihood of correctly ranking selection candidates was higher. The inbreeding problem detected in two of the three genotyped provenances could potentially be broken in the next generation due to mating among the genetically differentiated provenances.
Methods
Description of breeding seedling orchards (BSOs) and genetic material
The C. africana BSOs in this study were established at three sites in Ethiopia: Sekela Mariam Forest (SM), International Livestock Research Institute Ethiopia campus (ILRI), and Menagesha Suba Forest (Suba), as part of PATSPO, a national tree seed project in Ethiopia. These BSOs serve as test sites for C. africana germplasm, with the objective of developing an improved seed source through selection. The field experimental design for all locations was a randomised block design with single-tree plots.
The genetic material used in this study was obtained from 63 open-pollinated families from three native stands/provenances in Ethiopia: North Bench (P30), Adwa (P31), and Harar (P34). The number of families by provenance ranged from 20 to 23. Furthermore, bulk collections from 25 provenances lacking family information were also included. Details of the BSOs are summarised in Table 5.
Phenotyping and leaf sample collection
The height data (in decimetres) were collected from the three BSOs two years after planting. Families with high survival rate allowed for the sampling of an adequate number of individuals within the family, while also representing three main provenances in the country; thus, they were selected for SNP genotyping. Supplementary Table S3 provides details on the number of trees with phenotypic data and the number of trees sampled per genotyped family in each provenance. Leaf samples were collected from the ILRI BSO, silica gel dried in zip-lock bags for DNA extraction.
DNA extraction, genotyping, and data preprocessing
DNA was extracted with the DNeasy Plant Mini Kit from QIAGEN (Germany). The DNA samples were sent to Diversity Arrays Technology (DArT) (https://www.diversityarrays.com) in Canberra, Australia, for DArTSeq genotyping.
The DArTseq genotyping [69] resulted in 9591 raw SNPs in the 550 genotyped individuals. Missing data across each locus were calculated, and loci with missing data in 15% of the trees/individuals were excluded. The heterozygosity per locus was used as a further filtering criterion, in which loci with ≤ 0.05 heterozygosity were removed. The filtering of data was also done across samples, and individuals with missing data in 15% of the loci were excluded. These filtrations reduced the SNPs and individuals to 4373 and 526, respectively. Filtering was done using the qc.filtering function in the R package ASRgenomics [70].
Pedigree correction
Using the filtered SNPs, we validated and corrected the pedigree of the open-pollinated families based on a comparison of the expected versus observed additive genetic relationships. From the G-matrix estimated following VanRaden [29], we examined the samples’ pairwise additive relationship coefficients for large deviations from their expected values, and the correct mother was manually reassigned. Thirty-six trees were removed for parent conflict. Of the remaining 490 trees, the pedigree records were corrected for 15 trees. The final set of genotyped individuals ranged from 7 to 32 trees per family. Principal component analysis of the G-matrix was done to reveal potential grouping of the genotyped provenances. Supplementary Table S4 displays the number of trees analysed after correcting their pedigree and excluding trees with conflicting family information. Overall, 8070 trees were used for the analysis.
Statistical analysis
Due to spatial heterogeneity within the BSOs, and for computational efficiency, the statistical analysis was conducted in two stages following Cappa et al. [39]. First, single-site analyses were made using a pedigree-based classical a priori design model, and an a posteriori spatial model with a first-order autoregressive error (co)variance structure (AR1 × AR1) [36]. The following single-trait single-site pedigree-based individual-tree mixed model was used:
where \({\varvec{y}}\) is the vector of phenotypic data; \({\varvec{\beta}}\) is the vector of fixed effects for blocks and provenances; \({\varvec{b}}\) is the vector of random bulk seed lot effects; \({\varvec{a}}\) is the vector of random effects that represents additive genetic effects (or breeding values), following a normal distribution with zero mean and covariance matrix A \({\sigma }_{a}^{2}\), where A is the average numerator relationship matrix derived from the pedigree information [71] and \({\sigma }_{a}^{2}\) is the additive genetic variance; and \({\varvec{e}}\) is the vector of the random residual effects following a normal distribution with zero mean and (co)variance matrix \({{\varvec{I}}\sigma }_{e}^{2}\), where \({\varvec{I}}\) is the identity matrix and \({\sigma }_{e}^{2}\) is the residual error variance. For the spatial autoregressive model, vector \({\varvec{e}}\) was partitioned into spatially dependent (ξ) and spatially independent (η) residuals. Therefore, the residual (co)variance matrix can be expressed as \({\sigma }_{\upxi }^{2}\left[AR1\left({\uprho }_{col}\right) \otimes AR1\left({\uprho }_{row}\right)\right]+ {\sigma }_{\upeta }^{2}{\varvec{I}}\), where \({\sigma }_{\upxi }^{2}\) is the spatially dependent variance; \({\sigma }_{\upeta }^{2}\) is the spatially independent variance; \(AR1\)(ρ) is the first-order autoregressive correlation process; \({\uprho }_{col}\) and \({\uprho }_{row}\) are autocorrelations parameters for columns and rows, respectively; and ⊗ denotes the Kronecker product. \({\varvec{X}}\), \({{\varvec{Z}}}_{b}\), and \({{\varvec{Z}}}_{a}\), are incidence matrices relating fixed and random effects to measurements in vector \({\varvec{y}}\).
For individuals from bulk seed lots, an extra independent variance was fitted. This approach was used to avoid bias in estimates of additive genetic variances by the bulk trees, as they were considered unrelated when their actual relationship was unknown [72].
Finally, the adjusted phenotype data were generated by subtracting the estimated block and the autoregressive residual effects from the corresponding raw phenotypes.
In the second stage, the adjusted phenotypes were analysed using the following pedigree-based (ABLUP) multiple-site individual-tree mixed model:
where \({\varvec{y}}=\boldsymbol{ }\left[{{\varvec{y}}}_{1}^{\mathrm{^{\prime}}},{{\varvec{y}}}_{2}^{\mathrm{^{\prime}}},{{\varvec{y}}}_{3}^{\mathrm{^{\prime}}}\right]\) is the vector of individual tree adjusted-phenotypes for the sites (1 = ILRI, 2 = SM, and 3 = Suba); \({\varvec{\upbeta}}= \left[{{\varvec{\upbeta}}}_{1}^{\mathrm{^{\prime}}}, {{{\varvec{\upbeta}}}_{2}^{\mathrm{^{\prime}}},{\varvec{\upbeta}}}_{3}^{\mathrm{^{\prime}}}\right]\) is the vector of fixed effects of provenance for each site; \({\varvec{b}}= \left[{{\varvec{b}}}_{1}^{\mathrm{^{\prime}}}, {{{\varvec{b}}}_{2}^{\mathrm{^{\prime}}},{\varvec{b}}}_{3}^{\mathrm{^{\prime}}}\right]\) is the vector of random bulk seed lot effects for each site distributed as \({\varvec{b}}\sim {\varvec{N}}\left(0,{\boldsymbol{\Sigma }}_{{\varvec{b}}}\otimes{\varvec{I}}\right)\), where \({\boldsymbol{\Sigma }}_{{\varvec{b}}}\) is the (co)variance of the bulk seed lot effects; and \({\varvec{a}}= \left[{{\varvec{a}}}_{1}^{\mathrm{^{\prime}}}, {{{\varvec{a}}}_{2}^{\mathrm{^{\prime}}},{\varvec{a}}}_{3}^{\mathrm{^{\prime}}}\right]\) is the vector of additive genetic effects random vector distributed as \({\varvec{a}}\sim {\varvec{N}}\left(0,{\boldsymbol{\Sigma }}_{{\varvec{a}}}\otimes{\varvec{A}}\right)\), where \({\boldsymbol{\Sigma }}_{{\varvec{a}}}\) is the unstructured genetic effects (co)variance matrix between sites and \({\varvec{A}}\) is defined above;. Finally, \({\varvec{e}}= \left[{{\varvec{e}}}_{1}^{\mathrm{^{\prime}}}, {{\varvec{e}}}_{2}^{\mathrm{^{\prime}}}, {{\varvec{e}}}_{3}^{\mathrm{^{\prime}}}\right]\) is the vector of random residuals distributed as \({\varvec{e}}\sim {\varvec{N}}(0,{{\varvec{R}}}_{0}\otimes{\varvec{I}})\), where \({{\varvec{R}}}_{0}\) is the residual (co)variance matrix for the three sites with dimension 3 × 3. We assumed an unstructured (co)variance matrix for the genetic and bulk seed lot effects (\({\boldsymbol{\Sigma }}_{{\varvec{a}}}\) and \({\boldsymbol{\Sigma }}_{{\varvec{b}}}\), respectively). The matrices \({{\varvec{X}}}_{1}\), \({{\varvec{X}}}_{2}\), and \({{\varvec{X}}}_{3}\) and \({{\varvec{Z}}}_{{a}_{1}}\), \({{\varvec{Z}}}_{{a}_{2}}\), and \({{\varvec{Z}}}_{{a}_{3}}\) relate the observation to the means of the provenance effects in \({\varvec{\upbeta}}\) and the additive genetic effects for each tree in \({\varvec{a}}\). The symbol “´”, indicates the transpose operation.
The average-numerator relationship A-matrix was computed using the corrected pedigree data. In addition, a modified numerator relationship matrix (A2) that considers partial selfing was computed according to Dutkowski et al. [73]. Both matrices were created in the ASReml-R version 4.0 [74]. The ainverse function and the argument selfing were used to incorporate the selfing rate (s) estimated from the genotyped individuals (average s = 0.30; see results below). The mixed model that utilised the A-matrix was referred to as ABLUP-1, while the mixed model that used matrix A2 was referred to as ABLUP-2.
In the genomic-based GBLUP approach, the average numerator relationship matrix A (A-matrix) derived from pedigree information in the previous mixed models (2) was substituted by the genomic relationship matrix (G-matrix), estimated according to VanRaden [29] and based on the 4373 SNPs:
where \({\varvec{W}}\) is a matrix of order n x m (n = number of individuals, m = number of SNPs) with entries equal to wij = gij − 2pi, in which gij is the gene content at SNP locus i for tree j, and pi is the current allele frequency for marker i.
Finally, in the ssGBLUP approach, the A-matrix was substituted by the combined additive relationship H-matrix [30; 32], resulting from combining the G- with the A- or A2-matrix depending on whether inbreeding was accounted for:
where \({{\varvec{A}}}_{11}\) is the relationship matrix for non-genotyped individuals, \({{\varvec{A}}}_{22}\) is the pedigree-based relationship matrix for genotyped individuals, and \({{\varvec{A}}}_{12}\) and \({{\varvec{A}}}_{21}\) are the additive genetic relationship matrices between genotyped and non-genotyped individuals, respectively. \({{\varvec{G}}}_{{\varvec{w}}}\) is the marker-based relationship matrix for genotyped individuals weighted as: \({{\varvec{G}}}_{{\varvec{w}}}=0.90 {\varvec{G}}+0.10\boldsymbol{ }{{\varvec{A}}}_{22}\). The A-matrix was combined with the G-matrix to obtain the H-matrix, while matrix A2 was combined with the G-matrix to obtain the H2-matrix. The mixed model that utilised the H-matrix was referred to as ssGBLUP-1, while the mixed model that used the H2-matrix was referred to as ssGBLUP-2. The combined H-matrix for the ssGBLUP analysis was obtained using the R package ASRgenomics [70].
Genetic parameters
Restricted maximum likelihood [75] was used to estimate the (co)variances for the random effects in mixed models (1) and (2), and were obtained with the ASReml-R programme [74], which used the average information algorithm [76].
The narrow-sense individual heritability (\({h}^{2})\) and the additive genetic correlation (\({r}_{a}\)) between the three sites were estimated as:
where \({\sigma }_{a}^{2}\) is the additive genetic variance; \({\sigma }_{b}^{2}\) bulk (seed-lot) variance and \({\sigma }_{e}^{2}\) are the residual variances for each site; \({\sigma }_{{a}_{ij}}\) is the additive genetic covariance between sites i and j; and \({\sigma }_{{a}_{ii}}^{2}\) and \({\sigma }_{{a}_{jj}}^{2}\) are additive genetic variances for sites i and j, respectively, using a multi-environment model (2).
Theoretical accuracy of breeding values and efficiency of ssGBLUP
The theoretical accuracy (\(Acc\)) of the predicted breeding values was estimated using the following expression:
Where \(PEV\) is the prediction error variance and is calculated as the square of the standard error; \({F}_{i}\) is the inbreeding coefficients of the ith tree; and \({\sigma }_{a}^{2}\) is the additive genetic variance.
Efficiency (\(E\)) of ssGBLUP, which is the proportion of the extra benefit of GBLUP over ABLUP in the ssGBLUP scenario, was estimated following the method described in Sanchez-Mayor et al. [41].
When \(E\) = 1, ssGBLUP has the same performance as GBLUP. However, when \(E\) = 0, ssGBLUP has no advantage over ABLUP. This \(E\) parameter was calculated for the theoretical accuracy of the predicted breeding values.
Finally, the expected genetic gain, i.e., the change in the average breeding value of a population after selection, was calculated as follows: 1) the net breeding value of a tree was calculated as the sum of the estimated provenance fixed effect and the predicted breeding value of the tree within the provenance [77] from Eq. (2) of the ABLUP and ssGBLUP models with and without inbreeding; 2) the average of this net breeding value of the entire population was subtracted from the average net breeding value of the top 10% of selected trees (reported as a percent increase in average breeding value).
Predictive accuracy, prediction bias, and cross-validation scenarios
The predictive accuracy (PA) and prediction bias (PB) of all five models were evaluated using tenfold cross-validation, where 10% was used as the validation set and the remaining 90% as the training set. Two scenarios were tested to investigate the impact of provenance/population structure on PA and PB: 1) random sampling, where all measured trees were in the training population at least once; and 2) random sampling within the three provenances with strong genetic structure. The PA was determined as the Pearson correlation between the breeding values calculated from the full dataset and the ssGBLUP model (i.e., using marker and phenotype data of all the trees) and those predicted from the validation set using the ABLUP-1, ABLUP-2, ssGBLUP-1, ssGBLUP-2, and GBLUP models. PB was estimated by measuring the slope of the regression coefficient between the breeding values from the full dataset and the ssGBLUP model and those predicted with either the ABLUP-1, ABLUP-2, ssGBLUP-1, ssGBLUP-2, or GBLUP model. A regression coefficient equal to one means absence of bias.
Availability of data and materials
Our field studies and experimental research on plants, as well as the collection of plant material, adhered to all institutional, national, and international guidelines and legislation. Voucher specimens were not obtained for the trees that were sampled and described in the manuscript. Each tree was labeled with unique identifier (from nursery to field), ensuring the preservation of their identity throughout genomic and phenotypic measurements.
The phenotype, pedigree, and genotype data generated and/or analyzed during the current study are publicly available in the ERDA repository: Public Archive: https://doi.org/10.17894/ucph.319eafee-91a8-4405-9242-78c85f0b5d14.
Abbreviations
- ABLUP:
-
Pedigree-based Best Linear Unbiased Prediction
- ABLUP-1:
-
ABLUP model without accounting for inbreeding
- ABLUP-2:
-
ABLUP model accounting for inbreeding
- BLUE:
-
Best Linear Unbiased Estimation
- BSO:
-
Breeding Seedling Orchard
- PATSPO:
-
Provision of Adequate Tree Seed Portfolio
- DArT:
-
Diversity Arrays Technology
- GBLUP:
-
Genomic Best Linear Unbiased Prediction
- ILRI:
-
International Livestock Research Institute
- PA:
-
Predictive Accuracy
- PB:
-
Prediction Bias
- SM:
-
Sekela Mariam
- ssGBLUP:
-
Single-step Genomic Best Linear Unbiased Prediction
- ssGBLUP-1:
-
ssGBLUP model without accounting for inbreeding
- ssGBLUP-2:
-
ssGBLUP model accounting for inbreeding
- Suba:
-
Menagesha Suba
References
König LA, Medina-Vega JA, Longo RM, Zuidema PA, Jakovac CC. Restoration success in former Amazonian mines is driven by soil amendment and forest proximity. Philos Trans R Soc B. 2023;378(1867). https://doi.org/10.1098/rstb.2021.0086
Stanturf JA, Madsen P, Sagheb-Talebi K, Hansen OK. Transformational restoration: Novel ecosystems in Denmark. Plant Biosyst. 2018;152:536–46.
Paul C, Brandl S, Friedrich S, Falk W, Härtl F, Knoke T. Climate change and mixed forests: how do altered survival probabilities impact economically desirable species proportions of Norway spruce and European beech?. Ann For Sci. 2019;76(14). https://doi.org/10.1007/s13595-018-0793-8
Le HD, Smith C, Herbohn J, Nguyen H. A Comparison of Growth, Structure and Diversity of Mixed Species and Monoculture Reforestation Systems in the Philippines. J Sustain Forest. 2020;40(4):401–30. https://doi.org/10.1080/10549811.2020.1767145.
Shaw PD, Graham M, Kennedy J, Milne I, Marshall DF. Helium: visualization of large scale plant pedigrees. BMC Bioinform. 2014;15:259. https://doi.org/10.1186/1471-2105-15-259.
Falconer DS, Mackay TFC. An Introduction to Quantitative Genetics. 4th ed. London: Prentice Hall; 1996.
Frankham R, Ballou JD, Briscoe DA. Introduction to conservation genetics. New York, NY: Cambridge University Press. 2002. https://doi.org/10.1017/CBO9780511808999
Ellstrand NC. Multiple paternity within the fruits of the wild radish. Raphanus sativus Am Nat. 1984;123:819–28. https://doi.org/10.1086/284241.
Gowaty PA, Karlin AA. Multiple maternity and paternity in single broods of apparently monogamous eastern bluebirds (Sialia sialis). Behav Ecol Sociobiol. 1984;15:91–5. https://doi.org/10.1007/BF00299374.
Meagher TR, Thompson EA. The relationship between single and parent pair genetic likelihoods in genealogy reconstruction. Theor Popul Biol. 1986;29:87–106.
Thompson EA. The estimation of pairwise relationships. Ann Hum Genet. 1975;39:173–88. https://doi.org/10.1111/j.1469-1809.1975.tb00120.x.
Lambeth C, Lee BC, O’Malley D, Wheeler N. Polymix breeding with parental analysis of progeny: an alternative to full-sib breeding and testing. Theor Appl Genet. 2001;103:930–43.
Grattapaglia D, Ribeiro VJ, Rezende GDSP. Retrospective selection of elite parent trees using paternity testing with microsatellite markers: an alternative short-term breeding tactic for Eucalyptus. Theor Appl Genet. 2004;109:192–9.
El-Kassaby YA, Lstiburek M. Breeding without breeding. Genet Res. 2009;91(2):111–20. https://doi.org/10.1017/S001667230900007X.
Hansen OK, McKinney LV. Establishment of a quasi-field trial in Abies nordmanniana — test of a new approach to forest tree breeding. Tree Genet Genomes. 2010;6(2):345–55. https://doi.org/10.1007/s11295-009-0253-6.
Aykanat T, Johnston SE, Cotter D, Cross TF, Poole R, Prodőhl PA, et al. Molecular pedigree reconstruction and estimation of evolutionary parameters in a wild Atlantic salmon river system with incomplete sampling: a power analysis. BMC Evol Biol. 2014;14:68. https://doi.org/10.1186/1471-2148-14-68.
Askew GR, El-Kassaby YA. Estimation of relationship coefficients among progeny derived from wind-pollinated orchard seeds. Theor Appl Genet. 1994;88:267–72.
Namkoong G, Kang HC, Brouard JS. Tree breeding: principles and strategies. Theo Appl Genet Mono 1988;11. https://doi.org/10.1007/978-1-4612-3892-8
Vidal M, Plomion C, Harvengt L, Raffin BC, Bouffier L. Paternity recovery in two Maritime pine polycross mating designs and consequences for breeding. Tree Genet & Genomes. 2015;11:105. https://doi.org/10.1007/s11295-015-0932-4.
Tambarussi EV, Pereira FB, da Silva PHM, Lee D, Bush M. Are tree breeders properly predicting genetic gain? A case study involving Corymbia species. Euphytica. 2018;214:150. https://doi.org/10.1007/s10681-018-2229-9.
Klapste J, Suontama M, Dungey H, Telfer E, Graham N, Low C, et al. Effect of hidden relatedness on single-step genetic evaluation in an advanced open-pollinated breeding program. J Hered. 2018;109(7):802–10. https://doi.org/10.1093/jhered/esy051.
Mondini L, Noorani A, Pagnotta MA. Assessing plant genetic diversity by molecular tools. Diversity. 2009;1(1):19–35.
Alemayehu G, Asfaw Z, Kelbessa E. Cordia africana (Boraginaceae) in Ethiopia: A review on its taxonomy, distribution, ethnobotany and conservation status. Int J Botany Stud. 2016;1(2):38–46.
Wassie A. Opportunities, constraints and prospects of Ethiopian Orthodox Tewahdo Church in conserving forest resources. M.Sc: Thesis, Swedish University of Agricultural Sciences; 2004.
Derero A, Gailing O, Finkeldey R. Maintenance of genetic diversity in Cordia africana Lam., a declining forest tree species in Ethiopia. Tree Genet Genomes 2011;7:1–9. https://doi.org/10.1007/s11295-010-0310-1
Regassa R. Diversity and conservation status of some economically valued indigenous medicinal plants in Hawassa College of Teacher Education Campus. Southern Ethiopia Int J Adv Res. 2013;1(3):308–28.
PATSPO (https://www.worldagroforestry.org/project/provision-adequate-tree-seed-portfolio-ethiopia).
Veerkamp RF, Mulder HA, Thompson R, Calus MP. Genomic and pedigree-based genetic parameters for scarcely recorded traits when some animals are genotyped. J Dairy Sci. 2011;94(8):4189–97. https://doi.org/10.3168/jds.2011-4223.
VanRaden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91:4414–23.
Legarra A, Aguilar I, Misztal I. A relationship matrix including full pedigree and genomic information. J Dairy Sci. 2009;92:4656–63.
Aguilar I, Misztal I, Johnson DL, Legarra A, Tsuruta S, Lawlor TJ. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J Dairy Sci. 2010;93(2):743–52. https://doi.org/10.3168/jds.2009-2730.
Christensen OF, Lund MS. Genomic prediction when some animals are not genotyped. Genet Sel Evol. 2010;42:2.
Ratcliffe B, Gamal El-Dien O, Cappa EP, Porth I, Klapste J, Chen C, et al. Single step BLUP with varying genotyping effort in open-pollinated Picea glauca. G3-Genes Genom Genet. 2017;7:935–42.
Cappa EP, El-Kassaby YA, Muñoz F, Garcia MN, Villalba PV, Klápště, J et al. Genomic-based multiple-trait evaluation in Eucalyptus grandis using dominant DArT markers. Plant Sci. 2018;271:27–33. https://doi.org/10.1016/j.plantsci.2018.03.014
Callister AN, Bradshaw BP, Elms S, Gillies RAW, Sasse JM and Brawner JT. Single-step genomic BLUP enables joint analysis of disconnected breeding programs: an example with Eucalyptus globulus Labill. G3 (Bethesda) 2021;27:11(10):jkab253. https://doi.org/10.1093/g3journal/jkab253. PMID: 34568915
Cappa EP, de Lima BM, da Silva-Junior OB, Garcia CC, Mansfield SD, Grattapaglia D. Improving genomic prediction of growth and wood traits in Eucalyptus using phenotypes from non-genotyped trees by single-step GBLUP. Plant Sci. 2019;284:9–15. https://doi.org/10.1016/j.plantsci.2019.03.017.
Thavamanikumar S, Arnold RJ, Luo J, Thumma BR. Genomic studies reveal substantial dominant effects and improved genomic predictions in an open-pollinated breeding population of Eucalyptus pellita. G3-Genes Genom Genet. 2020;10(10):3751–3763. https://doi.org/10.1534/g3.120.401601
Ukrainetz NK, Mansfield SD. Prediction accuracy of single-step BLUP for growth and wood quality traits in the lodgepole pine breeding program in British Columbia. Tree Genet Genomes. 2020;16:64. https://doi.org/10.1007/s11295-020-01456-w.
Cappa EP, Ratcliffe B, Chen C, Thomas BR, Liu Y, Klutsch J, et al. Improving lodgepole pine genomic evaluation using spatial correlation structure and SNP selection with single-step GBLUP. Hered. 2022;128:209–24. https://doi.org/10.1038/s41437-022-00508-2.
Legarra A, Aguilar I, Colleau JJ. Short communication: Methods to compute genomic inbreeding for ungenotyped individuals. J Dairy Sci. 2020;103:3363–7. https://doi.org/10.3168/jds.2019-17750.
Sanchez-Mayor M, Riggio V, Navarro P, Gutiérrez-Gil B, Haley CS, De la Fuente LF, et al. Effect of genotyping strategies on the sustained benefit of single-step genomic BLUP over multiple generations. Genet Sel Evol. 2022;54(1):1–14. https://doi.org/10.1186/s12711-022-00712-y.
Ray D, Berlin M, Alia R, Sanchez L, Hynynen J, González-Martinez S, et al. Transformative changes in tree breeding for resilient forest restoration. Front For Glob Change. 2022;5:1005761. https://doi.org/10.3389/ffgc.2022.1005761.
White TL, Adams WT, Neale DB. Forest Genetics. Cambridge, MA: CABI Publishing; 2017. p. 682.
Grattapaglia D, Silva-Junior OB, Resende RT, Cappa EP, Müller BSF, Tan B, et al. Quantitative genetics and genomics converge to accelerate forest tree breeding. Front Plant Sci. 2018;871:1–10. https://doi.org/10.3389/fpls.2018.01693.
Hodge GR, Volker PW, Potts BM, Owen JV. A comparison of genetic information from open-pollinated and control-pollinated progeny tests in two eucalypt species. Theor Appl Genet. 1996;92:53–63.
Gamal El-Dien O, Ratcliffe B, Klápště J, Porth I, Chen C, El-Kassaby YA. Implementation of the Realized Genomic Relationship Matrix to Open-Pollinated White Spruce Family Testing for Disentangling Additive from Non additive Genetic Effects. G3; Genes Genom Genet. 2016;6:743–753. https://doi.org/10.1534/g3.115.025957PMID:26801647
Tan B, Grattapaglia D, Martins GS, Ferreira KZ, Sundberg B, Ingvarsson PK. Evaluating the accuracy of genomic prediction of growth and wood traits in two Eucalyptus species and their F1 hybrids. BMC Plant Biol. 2017;17:110. https://doi.org/10.1186/s12870-017-1059-6. (PMID:28662679).
Quezada M, Aguilar I, Balmelli G. Genomic breeding values’ prediction including populational selfing rate in an open-pollinated Eucalyptus globulus breeding population. Tree Genet Genomes. 2022;18:10. https://doi.org/10.1007/s11295-021-01534-7.
Cappa EP, Klutsch JG, Sebastian-Azcona J, Ratcliffe B, Wei XJ, Da Ros L, et al. Integrating genomic information and productivity and climate-adaptability traits into a regional white spruce breeding program. PLoS ONE. 2022;17:e0264549. https://doi.org/10.1371/journal.pone.0264549.
Walker TD, Cumbie WP, Isik F. Single-Step genomic analysis increases the accuracy of within-family selection in a clonally replicated population of Pinus taeda L. For Sci. 2022;68:37–52.
Gamal El-Dien O, Ratcliffe B, Klápště J, Chen C, Porth I, El-Kassaby YA. Prediction accuracies for growth and wood attributes of interior spruce in space using genotyping-by-sequencing. BMC genom. 2015;16(1):1–16. https://doi.org/10.1186/s12864-015-1597-y. (PMID:25956247).
Lenz PRN, Beaulieu J, Mansfield SD, Cle ́ment S, Desponts M, Bousquet J. Factors affecting the accuracy of genomic selection for growth and wood quality traits in an advanced breeding population of black spruce (Picea mariana). BMC Genom. 2017;18:335 https://doi.org/10.1186/s12864-017-3715-5PMID:28454519
Chen ZQ, Baison J, Pan J, Karlsson B, Andersson B, Westin J, et al. Accuracy of genomic selection for growth and wood quality traits in two control-pollinated progeny trials using exome capture as the genotyping platform in Norway spruce. BMC genom. 2018;19(1):1–16. https://doi.org/10.1186/s12864-018-5256-y.
Legarra A. Comparing estimates of genetic variance across different relationship models. Theor Popul Biol. 2016;107:26–30. https://doi.org/10.1016/j.tpb.2015.08.005. (PMID:26341159).
Cappa EP, El-Kassaby YA, Muñoz F, Garcia MN, Villalba PV, Klápště J, et al. Improving accuracy of breeding values by incorporating genomic information in spatial-competition mixed models. Mol Breed. 2017;37:1–13. https://doi.org/10.1007/s11032-017-0725-6.
Kainer D, Stone EA, Padovan A, Foley WJ, Külheim C. Accuracy of Genomic Prediction for Foliar Terpene Traits in Eucalyptus polybractea, G3-Genes Genom Genet. 2018;8(8):2573–2583. https://doi.org/10.1534/g3.118.200443
Habier D, Fernando RL, Dekkers JC. The impact of genetic relationship information on genome-assisted breeding values. Genet. 2007;177(4):2389–97. https://doi.org/10.1534/genetics.107.081190.
Habier D, Fernando RL, Garrick DJ. Genomic BLUP Decoded: A look into the black box of genomic prediction. Genet. 2013;194(3): 597607. https://doi.org/10.1534/genetics.113.152207.
Grattapaglia D, Resende MDV. Genomic selection in forest tree breeding. Tree Genet Genomes. 2011;7:241–55.
Calleja-Rodriguez A, Pan J, Funda T, Chen Z, Baison J, Isik F et al. Evaluation of the efficiency of genomic versus pedigree predictions for growth and wood quality traits in Scots pine. BMC Genom. 2020;21:1–17. https://doi.org/10.1186/s12864-020-07188-4
Isik F. Genomic Prediction of Complex Traits in Perennial Plants: A Case for Forest Trees. In: Ahmadi, N., Bartholomé, J. (eds) Genomic Prediction of Complex Traits. Methods in Molecular Biology, vol 2467. Humana, New York, NY. 2022. https://doi.org/10.1007/978-1-0716-2205-6_18
Crossa J, Pérez‐Rodríguez P, Cuevas J, Montesinos‐López O, Jarquín D, de los Campos, G et al. Genomic selection in plant breeding: Methods, models, and perspectives. Trends Plant Si. 2017;22(11):961–975. https://doi.org/10.1016/j.tplants.2017.08.011
Arojju SK, Conaghan P, Barth S, Milbourne D, Casler MD, Hodkinson TR, et al. Genomic prediction of crown rust resistance in Lolium perenne. BMC Genet. 2018;19:35. https://doi.org/10.1186/s12863-018-0613-z.
Rosvall O. Enhancing gain from long-term forest tree breeding while conserving genetic diversity. Umeå, Sweden: Swedish University of Agricultural Sciences; 1999.
Durel CE, Bertin P, Kremer A. Relationship between inbreeding depression and inbreeding coefficient in maritime pine (Pinus pinaster). Theor Appl Genet. 1996;92:347–56. https://doi.org/10.1007/BF00223678.
Bouffier L, Raffin A, Kremer A. Evolution of genetic variation for selected traits in successive breeding populations of maritime pine. Hered. 2008;101:156–65. https://doi.org/10.1038/hdy.2008.41.
Taylor PD, Fahrig L, Henein K, Merriam G. Connectivity is a vital element of landscape structure. Oikos. 1993;68:71–573.
White GM, Boshier DH, Powell W. Increased pollen flow counteracts fragmentation in a tropical dry forest: an example from Swietenia humilis Zuccarini. Proc Natl Acad Sci USA. 2002;99:2038–42.
DArTseq genotyping. https://www.diversityarrays.com/services/dartseq/dartseq-data-types/.
Gezan SA, de Oliveira AA, Murray D. ASRgenomics: An R package with Complementary Genomic Functions. Version 1.0.0 VSN International, Hemel Hempstead, United Kingdom. 2021.
Henderson CR. Applications of linear models in animal breeding. Guelph: University of Guelph; 1984.
Dutkowski GW, Silva JC, Gilmour AR, Wellendorf H, Aguiar A. Spatial analysis enhances modelling of a wide variety of traits in forest genetic trials. Can J For Res. 2006;36:1851–70. https://doi.org/10.1139/X06-059.
Dutkowski G, Gilmour A, Borralho N. Modification of the additive relationship matrix for open pollinated trial. In: Barros S, Ipinzà R, editors. Developing the eucalypt of the future. Proceedings of IUFRO Working Group 2.08.03 Conference, 10–15 September, Valdivia, Chile. 2001.
Butler DG, Cullis BR, Gilmour AR, Gogel BJ, Thompson R. ASReml-R reference manual version 4. 2018. http://www.homepages.ed.ac.uk/iwhite/asreml/uop
Patterson HD, Thompson R. Recovery of inter-block information when block sizes are unequal. Biometrika. 1971;58:545–54.
Gilmour AR, Thompson R, Cullis BR. Average information REML, an efficient algorithm for variance parameter estimation in linear mixed models. Biom J. 1995;51:1440–50.
Phocas F, Laloë D. Should genetic groups be fitted in BLUP evaluation? Practical answer for the French AI beef sire evaluation. Genet Sel Evol. 2004;36(3):325–45. https://doi.org/10.1186/1297-9686-36-3-325.
Acknowledgements
We thank PATSPO for financial support and for allowing us access to the BSOs and the PATSPO staff, namely senior team leader Søren Moestrup for supporting and facilitating the financing and field work, seed source expert Carsten Tom Nørgaard, and seed source officer Dr. Abrham Abiyu for phenotyping and help with sample collection, and field assistant Bekele Achame for helping with sample collection. We also thank lab technician Lene Hasmark Andersen and student assistant Camilla Frost Holm from IGN for sample preparation and DNA extraction.
Funding
Open access funding provided by Copenhagen University This project has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie Grant Agreement No 801199. The fieldwork, lab expenses and genotyping costs were covered by PATSPO.
Author information
Authors and Affiliations
Contributions
Conceptualisation: KMO, OKH and PH; Study design: KMO, EPC and OKH; Sample collection and DNA extraction: KMO; Data analysis: KMO with supervision from EPC, OKH and JKH; First draft: KMO; Methodological advice, Reviews, Comments and Edits of the text: All authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ousmael, K.M., Cappa, E.P., Hansen, J.K. et al. Genomic evaluation for breeding and genetic management in Cordia africana, a multipurpose tropical tree species. BMC Genomics 25, 9 (2024). https://doi.org/10.1186/s12864-023-09907-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-023-09907-z