Abstract
Background
Pancreatic ductal adenocarcinoma (PDAC) remains one of the most fatal malignancies. Several chemotherapies employing fluorouracil (5-FU) and gemcitabine were attempted, but the survival rate was extremely low. Resveratrol (RVT), known as a polyphenol compound and phytoalexin, was demonstrated to induce intrinsic apoptosis in cancer cells. However, its low delivery performance and efficiency at tumor sites remain an obstacle to exploit RVT as a drug. To address these problems, we bio-conjugated resveratrol with gold nanoparticles (GNPs) via polyvinylpyrrolidone as a cross-linker (RVT@PVP-GNPs) and investigated whether the fabrications could enhance the delivery performance and anti-tumor efficacy of RVT.
Results
The fabrication of gold nanoparticles (GNPs) and bio-conjugated with resveratrol (RVT@PVP-GNPs) was conducted firstly. TEM image, spectrophotometry and zeta-potential revealed that the GNPs and RVT@PVP-GNPs having a size of approximately 40 nm were successfully synthesized and exhibited moderate stability. GNPs alone represented no damage in PANC-1 cells and moreover diminished the cytotoxicity of RVT in Raw264.7 murine macrophage cells, demonstrating the superiority of gold nanoparticles as a drug carrier. Evaluation using dialysis showed a burst release rate of RVT within 96 h at pH 5.0, demonstrating the possibility of enhanced efficiency of RVT delivery through blood vessels to the tumor. The RVT@PVP-GNPs induced increased rates of S-phase cell cycle arrest and apoptosis compared with free RVT. Notably, RVT@PVP-GNPs diminished the proportion of necrotic cells, whereas free RVT increased it. We also demonstrated that the RVT@PVP-GNPs may induce an apoptosis via intrinsic mitochondria with higher degree compared with free RVT, indicating the possibility of enhanced anti-tumor agents. In animal studies, RVT@PVP-GNPs conjugated with AS1411 aptamer induced efficient tumor volume suppression without accumulation in or damage to the kidneys in vivo.
Conclusions
The results demonstrate that RVT@PVP-GNPs enhance the anti-tumor efficacy of free RVT by activating the intrinsic apoptotic pathway and could be considered as potential anti-tumor drug candidates against pancreatic cancer cells.
Similar content being viewed by others
Background
Pancreatic ductal adenocarcinoma (PDAC) was ranked third most common cause of cancer-associated mortality in the United States (Sung et al. 2021). Owing to poor prognosis, PDAC is considered one of the most fatal malignancies with a surviving 5 years of 10%. Surgical resection was the major treatment for PDAC but 5-year survival rate following surgery remained only 20% due to difficulties of diagnosis in early stage (Mizrah et al. 2020). Several chemotherapies employing fluorouracil (5-FU) and gemcitabine were also attempted up to date, but the efficacy was reduced by drug resistance or recurrence (Amrutkar et al. 2017). Therefore, identification of new therapeutic strategies for PDAC treatment is still needed.
One strategy for PDAC treatment is to exploit nano-carrier system offering novel attributes that include simple surface modification, enhanced internalization and delivery efficiency and target-specific delivery. These novel attributes made the systems highly suitable for drug delivery (Shi et al. 2017; Qiao et al. 2010; Germain et al. 2020). Diverse nanoparticles have been synthesized and modified for use as nano-carrier systems. Above all, gold nanoparticles (GNPs) have emerged as attractive candidates for enhanced drug delivery owing to their unique biological and chemical attributes, including targeted delivery, biocompatibility and lower toxicity than other metal nanoparticles (Ghosh et al. 2008; Elahi et al. 2018; Singh et al. 2018). Over the past decades, GNPs have been characterized and developed to deliver several therapeutic drugs, including nucleic acids, peptides, proteins, and natural compounds, by passive or active targeting (Kong et al. 2017; Chowdhury et al. 2017). More recently, thermosensitive GNPs have been designed for delivery to target sites for diagnosis by utilizing their highly efficient photo-thermal effects (Vines et al. 2019).
Several approaches have been attempted to synthesize GNPs including the Turkevich method (Turkevich et al. 1951). This method is known to synthesize GNPs easily and rapidly with the attachment of a few chemical compounds. However, the size of the synthesized GNPs varied markedly as gold ion reduction was conducted at a single step. Cellular uptake or internalization was disturbed due to the varying sizes of GNPs; therefore, therapeutic efficiency was decreased (Zhang et al. 2011). To reduce the size variation of GNPs, we used the seed-growth method in the present study, which could control the size of synthesized GNPs and enabled easy drug-loading (Lee et al. 2019).
Resveratrol (RVT), a polyphenol compound and phytoalexin, is secreted by several plants in self-defense against stress conditions or bacterial invasion (Ma et al. 2018). Many studies have previously reported its diverse benefits, including anti-inflammatory effects and cardiovascular system protection by lowering blood pressure (Kim et al. 2022; Bonnefont-Rousselot. 2016; He et al. 2010). RVT is also known to have anti-tumor effects in several cancers by apoptotic signaling pathway via mitochondria. A recent study demonstrated that the level of pro-apoptotic proteins and caspases were highly upregulated after RVT treatment (Liang et al. 2021). However, a major issue in utilizing polyphenol compounds in therapy is its low delivery performance and efficiency at tumor sites, highlighting the need of carriers as drugs (Gimenez-Bastida et al. 2019; Li et al. 2020).
Here, we designed RVT-loaded GNPs with polyvinylpyrrolidone (PVP) as a cross-linker (RVT@PVP-GNPs) and investigated whether the fabrications could enhance the delivery performance and anti-tumor efficacy of RVT. The fabrications were treated on PANC-1 human pancreatic cells and subsequently in vivo mouse model. Our findings also demonstrated novel potentiality of GNPs stability as drug carriers.
Results
Nanoparticle characterization
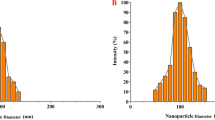
We hypothesized that resveratrol (RVT)-loaded gold nanoparticles (GNPs) could enhance the anti-tumor efficacy of RVT alone. To elucidate the hypothesis, we aimed to synthesize GNPs using the seed-growth method, which might be more suitable for generating nanoparticles of the desired size than the conventional Turkevich method (Zhang et al. 2011; Wuithschick et al. 2015). As expected, GNPs that were synthesized by seed-growth method showed a size of approximately 30–40 nm with low variation (Fig. 1A-a and b, 1B). They were then conjugated with RVT using polyvinylpyrrolidone (PVP). TEM images revealed that GNPs were surrounded by RVT through PVP (RVT@PVP-GNPs; Fig. 1A-c and d). For further characterization, we estimated the polydispersity (PDI) of GNPs and RVT@PVP-GNPs using Nanobrook 90 plus. The PDI values were found to be 0.319 ± 0.006 and 0.220 ± 0.009 for GNPs and RVT@PVP-GNPs, respectively (Fig. 1B). Zeta potential values of GNPs and RVT@PVP-GNPs were 33.34 ± 1.41 mV and 26.05 ± 1.06 mV, respectively, indicating moderate stability of the synthesized nanoparticles in solution (Ben Tahar et al. 2019). The absorbance spectra of PVP-RVT, PVP-GNPs, and RVT@PVP-GNPs were acquired using a UV/visible spectrophotometer (Fig. 1C). RVT was known to have a maximum absorption peak at approximately 320 nm, whereas GNPs demonstrated it at 530 nm (Jannin et al. 2004; Buso et al. 2008). As depicted in Fig. 1C, PVP-RVTs and PVP-GNPs showed maximum absorption peaks at approximately 320 nm and 530 nm, respectively; whereas, RVT@PVP-GNPs showed maximum peaks at both 320 nm and 530 nm, indicating the formation of RVT-loaded GNPs (Gangwar et al. 2012).
TEM and UV-spectrum of gold nanoparticles bio-conjugated with resveratrol (RVT@PVP-GNPs). A TEM images of synthesized gold nanoparticles (GNPs) and RVT@PVP-GNPs. a and b Represent gold nanoparticles (GNPs) with high or low magnification, respectively, whereas c and d represent RVT@PVP-GNPs with high or low magnification, respectively. The scale bar is indicated on the bottom left of each image. B Diameter, polydispersity (PDI), and zeta-potential values of synthesized GNPs and RVT@PVP-GNPs. C UV/visible spectra of RVT@PVP-GNPs (red), PVP-functionalized GNPs (blue), and resveratrol in PVP (green)
In vitro drug cumulative release
The release rate of RVT from RVT@PVP-GNPs was estimated at intervals for 96 h at two pH values (pH = 7.4 and 5.0) (Fig. 2). Tumor cell tissues deviated from the normal cell cycle; therefore, tumor cell growth accelerated, leading to hypoxia due to inadequate oxygen supply (Hockel and Vaupel 2001). Cancer cells were also known to acidify their environment (Wilson and Hay 2011). Thus, a major goal for nano-carrier utilization could be an enhanced drug delivery using a pH-sensitive drug carrier. The percentage of released RVT showed almost no difference at pH 7.4 (31.36%) and pH 5.0 (26.08%) for up to 9 h. Then, higher levels of released RVT were observed at pH 5.0 than at pH 7.4, and the difference in the extent of released RVT increased with time up to 96 h (from 31.36% to 48.90% at pH 7.4, and from 26.08% to 79.71% for pH 5.0). This suggests that RVT release from RVT@PVP-GNPs is enhanced by decreased pH around target tumors.
Cell viability and DAPI/PI staining during treatment
We determined inhibitory concentration (IC50) of RVT or RVT@PVP-GNPs for PANC-1 cells by treating different sample concentrations after 72 h incubation at 37 °C (Fig. 3A). Groups treated with RVT and RVT@PVP-GNPs exhibited significantly decreased cell viability in a dose-dependent manner. More importantly, the RVT@PVP-GNPs group demonstrated a more rapid decrease in cell viability than that by the RVT group. This result agreed with those reported in previous studies where the drugs loaded onto GNPs demonstrated enhanced internalization to the cell membrane (Li et al. 2009; Shah et al. 2011). These results suggest that RVT@PVP-GNPs are internalized into cells to a larger extent than free RVT and have a higher efficiency at a lower dose than free RVT.
Cell viability after treatment of RVT or RVT@PVP-GNPs. A PANC-1 cell viability compared with that of the control after RVT or RVT@PVP-GNP treatment for 72 h. Diverse concentrations of RVT or RVT@PVP-GNPs were treated in PANC-1 cells before counting. B DAPI and PI double staining after drug treatment for 6 h. Cell nucleus was stained with DAPI. Cells undergoing apoptosis or necrosis were visualized by PI staining and observed using a fluorescence microscope at ×200 magnification
To investigate cell viability after treatment, they were stained with DAPI or PI fluorescence dye and observed under a fluorescence microscope (Fig. 3B). The GNPs-treated group demonstrated no difference in viability compared with the control after 6 h of incubation. In contrast, a higher number of PI-stained dead cells (red) were observed in the RVT@PVP-GNPs-treated group than in the free RVT group. It is known that PI fluorescence dye intercalates to DNA through permeable cell membranes. Therefore, staining with PI alone could show the ability of cell death by drugs but cannot distinguish between apoptotic and necrotic cells (Dive et al. 1992). Thus, we subsequently conducted flow cytometry to establish whether cell death is caused by apoptosis or necrosis.
We also analyzed the lactate dehydrogenase (LDH) leakage levels in Raw264.7 murine macrophage cell line (Additional file 1: Fig. S1). LDH is an enzyme that catalyzes conversion of lactate to pyruvate in most cells. It easily leaks out of cells damaged by foreign factors; hence, leaked LDH extent in solution was regarded as evidence of cytotoxicity (Weyermann et al. 2005). A significantly lower LDH leakage level was observed in the two GNP-included groups compared with the control or free RVT-included group, demonstrating lower cytotoxicity of the synthesized GNPs compared with that of the control (Additional file 1: Fig. S1).
Flow cytometry analysis
PANC-1 cells were treated with GNPs, RVT, and RVT@PVP-GNPs at their IC50 values and incubated for 72 h. The GNPs-treated group did not represent significant difference in the cell cycle distribution compared with the control group (Fig. 4A, B). However, RVT- and RVT@PVP-GNPs-treated groups showed a different pattern of cell cycle arrest when compared with the control and GNP-treated group. The RVT-treated group demonstrated a significantly higher proportion of cells in the S-phase (37.72 ± 4.53% of GNP-treated group to 50.37 ± 2.49% of RVT-treated group) and a significantly lower cell proportion in the G2/M phase (15.41 ± 3.86% of GNP-treated group to 4.69 ± 1.51% of RVT-treated group), but much less difference was observed in the G0/G1 phase. The RVT@PVP-GNPs-treated group demonstrated an even higher proportion of cells in the S-phase than any other groups (37.72 ± 4.53% of GNP-treated group to 60.05 ± 2.83% of RVT@PVP-GNPs-treated group).
Induction of apoptosis in PANC-1 cells. A, B Change of PANC-1 cell cycle progression after drug treatment. The cells were fixed with ethanol after 72 h treatment and evaluated using flow cytometry. The percentage of cell populations in each phase is illustrated in B. C, D Apoptotic cell analysis after drug treatment. Cells were stained with Annexin V/PI and analyzed using flow cytometry. The percentage of cell populations is illustrated in D. Data are expressed as the mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with the control
We next quantitated the expression of cyclin D1 to elucidate the above results (Additional file 1: Fig. S2). As shown in Additional file 1: Fig. S2, cyclin D1 level was significantly downregulated in RVT- or RVT@PVP-GNPs-treated group, compared to the control group. Moreover, the RVT@PVP-GNPs-treated group had significantly lower cyclin D1 expression than the RVT-treated group. These results suggest that treatment with RVT and RVT@PVP-GNPs induces S-phase cell cycle arrest.
We also performed flow cytometry analysis of the cell population to determine the apoptotic effects of each treatment (Fig. 4C, D). The cells were stained with Annexin V/PI to analyze the apoptotic cell population after treatment with GNPs, RVT, or RVT@PVP-GNPs after 48 h incubation. Numbers of apoptotic cells between GNP-treated and the control group showed no significant difference. These results suggest that the synthesized GNPs act as drug carriers without influencing the population of apoptotic cells. The RVT-treated group showed significantly increased levels of both early apoptotic (3.62 ± 0.7% of the control to 8.05 ± 0.9% of RVT-treated group) and late apoptotic population (4.65 ± 0.9% of the control to 17.57 ± 0.3% of RVT-treated group) (Fig. 4D). The RVT-treated group also had a significantly increased extent of necrotic cell population (1.30 ± 0.6% of the control group to 18.14 ± 4.8% in the RVT-treated group) (Fig. 4C). RVT@PVP-GNPs-treated group also displayed a significantly increased extent of early apoptotic cell population (3.62 ± 0.7% of the control to 9.74 ± 2.2% of RVT@PVP-GNPs-treated group) and late apoptotic cell population (4.65 ± 0.9% of the control to 15.00 ± 1.1% of RVT@PVP-GNP-treated group) (Fig. 4D). These results demonstrate that the RVT induces cell death by apoptosis, and that the effect is enhanced by conjugating it with gold nanoparticles.
Quantitative analysis of mRNA expression
To elucidate previous results, we quantified the mRNA levels of several apoptosis-associated genes (Fig. 5). The expression of these genes was similar between the GNPs and the control group as expected. However, RVT or RVT@PVP-GNPs groups demonstrated significant higher levels of caspase-3, -7, and -9 compared with those in the control. More interestingly, caspase-7 and caspase-9 levels were significantly higher in the RVT@PVP-GNPs group than in the RVT group (Fig. 5A–C).
Quantitative analysis of mRNA expression after drug treatment. A–D The expression of genes related to intrinsic apoptotic pathways was measured using real-time PCR. Transcript levels were normalized to those of the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) housekeeping gene. Data are expressed as the mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with the control
Moreover, BAK expression was significantly higher in the RVT and RVT@PVP-GNPs groups than in the control and GNPs groups (Fig. 5D). Furthermore, the RVT@PVP-GNPs group had higher BAK expression than the RVT group. These results demonstrate that the RVT@PVP-GNP nano-carriers we fabricated strongly induce apoptosis through the intrinsic apoptosis pathway at the mRNA level.
Quantitative analysis of protein expression
We subsequently interrogated the immunoblot analysis to identify the anti-tumor effects of drugs could be expressed till protein levels in vitro (Fig. 6). As expected, caspase-3 and caspase-7 were upregulated in RVT and RVT@PVP-GNPs groups compared with that in the control. These results corroborate the results obtained for mRNA levels; however, there were no significant differences between the RVT and RVT@PVP-GNPs groups (Fig. 6B). To further analyze the apoptosis pathway, the expression levels of bcl-2 associated X protein (BAX) and Bcl-2 associated agonist of cell death (BAD) were also verified. p53 and poly(ADP-ribose) polymerase (PARP) expression was also highly upregulated in the RVT and RVT@PVP-GNPs groups (Fig. 6B). We observed that the expression levels of p53 and PARP in the RVT or RVT@PVP-GNPs groups were noticeably higher. Moreover, p53 and PARP levels in RVT@PVP-GNPs group were significantly higher than those in the RVT group, indicating that the delivery efficacy of RVT into cells may be enhanced by conjugation with GNPs. These results suggest that the fabricated RVT@PVP-GNPs enhance the apoptotic effect in protein levels compared to RVT alone.
Quantitative analysis of protein expression after drug treatment. The bands analyzed using western blotting are illustrated in A. The expression of proteins was normalized to that of β-actin and is presented in B. Data are expressed as the mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001 compared with the control
In vivo study
Subsequently, in vivo study was conducted using a BALB/c nu/nu mouse model to elucidate the anti-tumor effects of RVT@PVP-GNPs. The efficacy of delivering drugs through the active targeting strategy in nano-carrier systems remains unknown, as no other products have been circulated in the market. However, Amanda and Rachel summarized the current state in the clinical setting (Pearce and O’Reilly 2019); they described that the advantages of active targeting include enhanced cellular uptake and delivery efficacy. Moreover, Yoo et al. reported several advantageous effects of biological ligands including aptamers for active targeting in various cancer cell lines (Yoo et al. 2019). For application of the biological ligand, the AS1411 aptamer (Additional file 1: Table S1) was employed for conjugation on the surface of RVT@PVP-GNPs to enhance the delivery efficiency in in vivo experiments. The AS1411 aptamer targets nucleolin, which was abundantly expressed on the surface of cancer cells (Soundararajan et al. 2008).
PANC-1 cells were injected into the right flank of BALB/c nu/nu mice to develop a heterotrophic xenograft mouse model. Each group was treated with appropriate samples twice a week for 33 days. No significant changes in appearance were observed, with respect to body weight. During the experiment, the tumor volumes of the saline- or GNP-treated groups increased gradually up to approximately 240 mm3 and 220 mm3, respectively, suggesting that GNP treatment has almost no effect on tumor growth inhibition (Fig. 7A). However, the group treated with a low dose (20 mg/kg) of RVT@PVP-GNPs demonstrated a slower increase in tumor volume, which was approximately half the tumor volume of the control group on the last day (i.e., the tumor inhibition rate was approximately 50%; Fig. 7D). More interestingly, the group treated with a high dose (50 mg/kg) of RVT@PVP-GNPs had a dramatically lower tumor volume throughout the observation period. The tumor inhibition rate was approximately 80% compared with that of the control (Fig. 7E). The results demonstrate that RVT@PVP-GNPs diminish the tumor volume in a dose-dependent manner.
Anti-tumor efficiency of RVT or RVT@PVP-GNPs in vivo. Each mouse group was administered with PBS, bare-GNPs, 20 mg/kg RVT@PVP-GNPs, or 50 mg/kg RVT@PVP-GNPs via tail vein injections twice a week. A, B Tumor growth curves and body weights of mice were monitored for 33 days and are illustrated in A and B, respectively. C, D At the end of experiment, tumors were removed from mice and pictured or weighed for each group
At the end of experiment, all mice were euthanized, and their tumor tissues were removed and homogenized to extract proteins and quantify their levels (Fig. 8A, B). In all experiments, the GNP-treated group demonstrated no statistical difference compared with those of the control, whereas the groups treated with a low dose (20 mg/kg) or high dose (50 mg/kg) of RVT@PVP-GNPs displayed a dose-dependent increase in the fold change of proteins examined. Caspases-3/7 and PARP were located downstream of the apoptosis pathway (Boulares et al. 1999). Following treatment with low dose (20 mg/kg) of RVT@PVP-GNPs, the expression levels of caspases-3/7 and PARP increased by at least 3–22 times, compared to those in the control, and by at least 40–90 times, compared to those in the control, on treatment with high doses (50 mg/kg) of RVT@PVP-GNPs. The results might be correlated to the reduced tumor volume after treatment with RVT@PVP-GNPs. Additionally, the extent of BAX and BAD, which are related to the intrinsic apoptosis pathway, was also upregulated after treatment with RVT@PVP-GNPs (Singh et al. 2004). Of note, the extent of increase of BAX and BAD proteins levels was also much higher with a high dose (50 mg/kg) than with a low dose (20 mg/kg) of RVT@PVP-GNPs, as observed for the other three proteins (caspase-3/7 and PARP). These results suggest that RVT@PVP-GNPs induce PANC-1 cell apoptosis via the intrinsic apoptosis pathway.
Quantitative analysis of protein expression in tumor tissues. A, B Bands analyzed using western blotting are illustrated in A. The expression of proteins was normalized to that of β-actin and is presented in B. C Section images of tumor tissues and the kidneys after treatment with PBS, bare-GNPs, 20 mg/kg RVT@PVP-GNPs, or 50 mg/kg RVT@PVP-GNPs. Sections were visualized utilizing a AxioVert 200 M microscope (Carl Zeiss, Oberkochen, Germany) at ×200 magnification. Data are expressed as the mean ± SEM. *p < 0.05, **p < 0.01 and ***p < 0.001 compared with the control
H&E staining of tumor tissues was conducted to observe any histological changes after RVT@PVP-GNPs treatment (Fig. 8C). Compared with the control treated with phosphate-buffered saline (PBS), tumor tissues treated with RVT@PVP-GNPs had more vacuolation and reduced many tumor cells that had undergone karyolysis, whereas the bare-GNP-treated group showed almost no difference with the control (Wang et al. 2018). Sliced kidney sections were also examined to observe any remnant drugs that were accumulated. Images of kidney sections demonstrated little difference in glomerulus among the control, GNP-treated, and RVT@PVP-GNPs-treated groups, demonstrating that the synthesized samples caused no kidney damage (EL Barky et al. 2020). These results indicate that RVT@PVP-GNPs have potential therapeutic effects on in vivo tumor tissues related to PANC-1 or other pancreatic cancer cells.
Discussion
Nano-delivery system offers various drug candidates for efficient delivery to target sites. Resveratrol (RVT), a polyphenol compound and phytoalexin, was researched for a long time as candidates for cancer treatment. However, its low delivery performance and uptake efficiency through digestive system were major issues for utilization as drug candidates (Gimenez-Bastida et al. 2019; Li et al. 2020). Here, we fabricated gold nanoparticles (GNPs) bio-conjugated with RVT to enhance anti-tumor efficacy and evaluated its effects in pancreatic cancer cells.
The synthesized GNPs were aimed to be sized around 30–40 nm, since several studies reported that nanoparticles with sizes ranging from 25–50 nm demonstrated higher cellular uptake and endocytosis than those of other sizes (Yuan et al. 2010; Chithrani et al. 2006). Additionally, a study reported that curcumin was successfully conjugated with GNPs through hydrogen bonds between curcumin and GNPs (Gangwar et al. 2012). Since RVT is included in the same polyphenol category as curcumin, therefore, we presumed that it could also be bio-conjugated with GNPs.
Several studies have reported that RVT induces G0/G1- or S-phase cell cycle arrest in breast, prostate, or pancreatic cancer cell lines and which component of the cycle is arrested is still controversial (Roy et al. 2011; Kotha et al. 2006). In our research, RVT bio-conjugated with gold nanoparticles (RVT@PVP-GNPs) demonstrated S-phase cell cycle arrest among a significantly higher proportion of cells, compared with free RVT. To cross-check the results of flow cytometry analysis, we performed immunoblotting for assessing levels of cyclin D1, which has a vital role in cell cycle progression and proliferation (Yang et al. 2006). Cyclin D1 expression was known to be downregulated in cells that are in S-phase arrest, and the immunoblotting result was consistent with that of flow cytometry analysis (Zhang et al. 2019; Joe et al. 2002).
Interestingly, RVT@PVP-GNPs group demonstrated considerably much smaller necrotic cell populations compared with RVT group in the flow cytometry analysis (Fig. 2E). Apoptosis is programmed cell death, whereas necrosis is unregulated cell death that can induce pro-inflammatory responses due to the release of intracellular components (Kanduc et al. 2002). Necrosis has been demonstrated to promote the release of damage-associated molecular patterns (DAMPs) including uric acid, HMGB1, and IL-33 from cells, resulting in immune responses (Martin et al. 2016). In our study, the RVT@PVP-GNPs group showed much smaller necrotic cell populations while apoptotic cell populations were still maintained, demonstrating RVT@PVP-GNPs could be used as a potential therapeutic drug for treat pancreatic cancers.
In the molecular biological assays including real-time PCR and immunoblotting, we estimated levels of several mRNA or protein that are associated with intrinsic apoptosis to determine the exact signaling pathway activated by our fabrication. Caspase-3 and caspase-7 are executioner caspases, which is a caspase family of cysteine proteases located downstream of the apoptotic pathway (Boatright and Salvesen 2003). Several apoptotic signals from extrinsic or intrinsic apoptotic pathways were transferred to downstream apoptotic pathways. Executioner caspases were activated by the caspase cascade resulting in apoptosis by cleaving cellular components (Villa et al. 1997). Thus, increases in cleaved caspase-3 and caspase-7 levels are suggested as strong evidence of apoptosis.
Caspase-9, another family of cysteine proteases, is an initiator caspase associated with the intrinsic apoptosis pathway. It is activated by dimerization when it binds to the apoptosome, and it continuously cleaves caspase-3 and caspase-7 (Utaipan et al. 2020). The bcl-2 homologous antagonist killer (BAK) is a pro-apoptotic regulator located in the mitochondrial outer membrane to induce the release of cytochrome c to promote apoptosome-dependent caspase activations (Nechushtan et al. 2001). It was demonstrated that BAX, when located in the cytosol in the normal state undergoes a conformational shift and is transported to mitochondria in response to apoptotic signals. As a pro-apoptotic gene, BAX binds to BAK to form BAX/BAK oligomer, which allowed cytochrome c to escape into the cytoplasm and activate the apoptotic caspase cascade (Nechushtan et al. 2001). BAD formed heterodimers with anti-apoptotic proteins, such as B-cell lymphoma 2 (Bcl-2), to sustain apoptosis (Condorelli et al. 2001). Thus, activation of cleaved caspase-9 and mitochondrial proteins including BAX, BAK or BAD demonstrate that the drug induces apoptosis through intrinsic apoptotic pathway.
Along with them, p53 played essential roles in cancer as tumor suppressor, including preventing gene mutation, repairing DNA, holding cell cycle, and initiating apoptosis (Sengupta and Harris 2005; Haupt et al. 2003). Cells that receive apoptotic signals activated p53 phosphorylation to promote apoptosis with tight control (Haupt et al. 2003). PARP, which is located in the cell nucleus and involved in DNA repair, was another key factor in apoptosis (Ahn et al. 2022). In programmed cell death, PARP induced poly(ADP-ribose) (PAR) production, which stimulated apoptosis inducing factor (AIF) from mitochondria (Ahn et al. 2022). All our results of mRNA and protein assays including animal studies represented a higher level of apoptosis in RVT@PVP-GNPs-treated group compared with free RVT, demonstrating its potential as a therapeutic drug for cancer treatment.
Conclusions
In summary, we synthesized gold nanoparticles (GNPs), fabricated with resveratrol (RVT) via PVP as a cross-linker (RVT@PVP-GNPs) and investigated its anti-tumor performances. Synthesized GNPs and RVT@PVP-GNPs showed stable and homogeneous size around 40 nm, demonstrating higher cellular uptake and endocytosis than those at other sizes. RVT@PVP-GNPs also showed moderate stability and burst release rate of RVT, revealing the possibility of efficient delivery performance through the blood vessels to the tumor. GNPs alone represented no damage in PANC-1 cells, and moreover, reduced the cytotoxicity according to RVT in Raw264.7 murine macrophage cells, demonstrating the advantages of gold nanoparticles as a drug carrier. Flow cytometry analysis revealed that RVT@PVP-GNPs induced higher S-phase cell cycle arrest than free RVT. Notably, RVT@PVP-GNPs represented a much lower extent of necrosis than free RVT. We also demonstrated that the RVT@PVP-GNPs induce an apoptosis via intrinsic mitochondria with higher degree compared to free RVT, indicating the possibility of enhanced anti-tumor agents. The RVT@PVP-GNPs conjugated with AS1411 aptamer also demonstrated dose-dependent suppression of tumor volume without accumulation or kidney damage in vivo. These results demonstrate the therapeutic potential of RVT-loaded GNPs to achieve successful nanomedicine-based cancer therapy.
Methods
Materials
Tetrachloroauric acid trihydrate (HAuCl4 ∙ 3H2O, 99.9%), cetrimonium bromide (CTAB, > 99%), trisodium citrate dihydrate (C6H5Na3O7 ∙ 2H2O, > 99%), l-ascorbic acid (C6H8O6, 99%), PVP (M.W. 40,000), RVT (> 99%), dimethyl sulfoxide (> 99.7%), propidium iodide (PI, > 94%), 4,6-diamidino-2-phenylindole dihydrochloride (DAPI, > 98%), dithiothreitol (DTT, > 99%) and bovine serum albumin (BSA) were obtained from Sigma Aldrich (St. Louis, MO, USA). Tris(hydroxymethyl)aminomethane (Tris, > 99.5%) and glycine were purchased from BioShop (Burlington, Ontario, Canada). Sodium dodecyl sulfate was obtained from Affymetrix (Santa Clara, CA, USA). Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum, and phosphate-buffered saline (PBS) for cell culture were obtained from Welgene (Daegu, Korea). All antibodies utilized in this study were purchased from Cell Signaling Technology (Beverly, MA, USA). All other chemicals and reagents were of analytical grade.
Animals
BALB/c nu/nu mice (female, 5 weeks old) were obtained from Orient Bio (Seongnam, Korea), housed in the Gyerim Experimental Animal Resource Center of Korea University (Seoul, Korea), and acclimatized for 2 weeks before being segregated into experimental groups. The mice were housed under a 12-h light/dark cycle at a temperature of 23 ± 4 °C and 60 ± 5% humidity. All animals were provided ad libitum access to food and water. All animal experiments were conducted in accordance with the guidelines and permission of the Korea University Institutional Animal Care and Use Committee (KUIACUC-2017-93).
Nanoparticle synthesis
GNPs were synthesized using seed-growth method with slight modifications (Zhang et al. 2011). In brief, 20 mL of an aqueous solution comprising 2.5 × 10–4 M of HAuCl4 and trisodium citrate was mixed with gentle stirring. After stirring, 0.6 mL of ice-cold 0.1 M NaBH4 was incorporated to synthesize the seed solution. Simultaneously, 200 mL of an aqueous solution comprising 2.5 × 10–4 M HAuCl4 and 0.08 M CTAB was prepared as the growth solution and heated at 45 °C until the solution appeared clear orange. The seed solution (5 mL) and 0.1 M ascorbic acid were added together to 7.5 mL growth solution with gentle stirring. The stirring was continued for approximately 15 min until the solution turned pale red. The mixture was then used as another seed solution, and the entire procedure was repeated until the GNPs reached an appropriate size.
Conjugation for RVT@PVP-GNPs
RVT was loaded onto the surface of GNPs by utilizing PVP as a cross-linker. The synthesized GNPs were centrifuged thrice (4500×g, 30 min, 25 °C) to remove other ingredients, including CTAB. The GNPs were dispersed in 80 mL of distilled water, and mixed with 1.67 × 10–5 M PVP at 45 °C. After 40 min, the mixture was cooled, and 4.5 × 10–2 M of RVT was incorporated with stirring for 6 h at 45 °C. Subsequently, unloaded RVT was removed by centrifuging thrice (2000×g, 30 min, 25 °C) and then stored at 4 °C.
Characterizations
Fabrications were imaged using Transmission electron microscopy (TEM; FEI Tecnai 20 Hillsboro, OR, USA) to verify their synthesis (Al-Musawi et al. 2020; Khashan et al. 2020). The size, polydispersity, and zeta-potential of the synthesized nanoparticles were evaluated using Nanobrook 90 plus (Brookhaven, GA, USA). The absorbance of the synthesized nanoparticles was evaluated at 320 nm using a UV/visible spectrophotometer (Ultrospec 2100 pro, Amersham BioSciences, Buckinghamshire, UK) (Rashid et al. 2021; Jihad et al. 2021).
In vitro drug cumulative assay
The pH-dependent release of RVT-loaded RVT@PVP-GNPs was investigated using a dialysis method. Two milliliter of RVT@PVP-GNPs was placed in a Slide-A-Lyzer™ MINI Dialysis Device (10 K MWCO; Thermo Fisher Scientific, MA, USA) and subjected to dialysis against 1 × PBS (pH 7.4) or acetate buffer (pH 5.0) with gentle shaking. At specific time intervals, equal volumes of samples were collected from each buffers until 96 h and replaced with fresh buffer solution. The cumulative release of RVT was estimated using a spectrophotometer by measuring absorbance at 320 nm (Ultrospec 2100 pro).
Cell viability test
The PANC-1 human pancreatic cancer cell line was obtained from Korean Cell Line Bank (KCLB, Seoul, Korea). The 5 × 105 cells were seeded in 100-mm cell culture dishes. Cells were treated with RVT or RVT@PVP-GNPs at the desired concentrations for 72 h. Then, harvested cells were stained with 0.4% trypan blue solution (Gibco, Gaithersburg, MD, USA). Stained cells were counted using a hemocytometer (Paul Marienfeld, Lauda-Königshofen, Germany) and represented as a percentage of the control.
DAPI/PI cell staining
A density of 3 × 105 cells/well PANC-1 cells were seeded in 6-well plates and treated with desired concentration for 6 h. Then, the cells were washed with 1 × PBS twice and stained with 15 μg/mL PI solution (in PBS), followed by 30 min incubation in the dark. The cells were then immersed in 4% paraformaldehyde (in PBS) and 1 μg/mL DAPI solution (in PBS) and incubated for 30 min in the dark, followed by washing with 1 × PBS twice (Jabir et al. 2021). After fixation and staining, the cells were washed with distilled water twice and immersed in 1 mL distilled water. Prepared cells were then observed with an AxioVert 200 inverted fluorescence microscope (Carl Zeiss, Oberkochen, Germany).
Lactate dehydrogenase (LDH) leakage assay
Raw264.7 murine macrophage cell line was obtained from KCLB. A density of 5 × 105 cells/well was seeded in 6-well plates and treated with desired concentration for 72 h. Then, the supernatants were collected by centrifugation (1500×g, 5 min, 25 °C) to remove detached cells. The LDH leakage level was estimated using an LDH cytotoxicity detection kit (Takara, Tokyo, Japan) according to manufacturer’s instructions by measuring at 490 nm wavelength and represented as absorbance normalized to that of the control.
Flow cytometry analysis for determining cell cycle arrest
PANC-1 cells stored overnight at − 20 °C after fixed in ice-cold 100% ethanol. The fixed cells were centrifuged (250×g, 5 min, 4 °C) and washed with PBS twice a day after. Then, cells were resuspended in 100 µL of propidium iodide (PI) solution comprising 50 μg/mL PI, 1 mg/mL RNase and 0.1% Triton X-100. After 30 min in the dark, the amount of PI-labeled DNA was conducted at an excitation/emission wavelength of 488/610 nm through a FACSCalibur flow cytometer (BD Biosciences, NJ, USA) (Ibrahim et al. 2021). Data were analyzed with ModFit LT™ software (version 3.0, Verity Software House, Topsham, ME, USA).
Annexin V/PI double staining for determining apoptosis
FITC Annexin V apoptosis detection kit I (BD Biosciences) was employed to evaluate the apoptotic cell population. Harvested PANC-1 cells were centrifuged (250×g, 5 min, 25 °C) after washed with PBS twice. Then, the pellets were incubated with 5 μL Annexin V-FITC and/or PI for 15 min in the dark after re-suspended in 300 μL Annexin V binding buffer. The samples were analyzed through a FACSCalibur flow cytometer at excitation/emission wavelengths of 488/530 nm and 488/617 nm, respectively (Al-Salman et al. 2020). The data were estimated utilizing BD CellQuest Pro software (version 5.2.1, Becton Dickinson, Franklin Lakes, NJ, USA) and the apoptotic ratio was estimate.
Quantitative analysis of mRNA expression
Total RNA was extracted using the RNeasy mini kit (Qiagen, MD, USA) and was quantified utilizing Nano-drop (Thermo Fisher Scientific). The Reverse Transcription System (Promega, WI, USA) was utilized to synthesize the first-strand cDNA according to the manufacturer’s instructions. TaqMan PreAmp MasterMix (Thermo Fisher Scientific) and TaqMan probes (Thermo Fisher Scientific) were used to quantify relative gene expression. qPCR was subsequently conducted on a CFX-96 real-time PCR machine (Bio-Rad, CA, USA) with the following cycle conditions: initial denaturation for 10 min at 95 °C, followed by 40 cycles of denaturation for 15 s at 95 °C and annealing/elongation at 60 °C for 1 min. GAPDH (control) was used to normalize the relative gene expression employing the 2(−∆∆CT) method (Livak and Schmittgen 2001).
Quantitative analysis of protein expression
The cells were lysed in RIPA buffer and measured protein concentration using a BCA protein assay kit (Thermo Fisher Scientific). Proteins were segregated through electrophoresis with varying percentages of polyacrylamide depending on the proteins sizes. Proteins were transferred to a polyvinylidene fluoride membrane (0.45 μm; EMD Millipore, MA, USA), and blocked for 1 h. The membrane was then stored at 4 °C overnight with primary antibody using an orbital shaker (DS300, BMS, Seoul, Korea). After washing with 1 × Tris-buffered saline containing 0.1% Tween-20, the blots were incubated with a horseradish peroxidase-conjugated secondary antibody for 2 h. The blots were visualized with enhanced chemiluminescence western blotting detection reagents (Thermo Fisher Scientific) and BioRad XRS+ imaging system (Bio-Rad).
Aptamer conjugation
For the animal study, the 5-thiol-modified aptamer, AS1411, was obtained from Bioneer (Daejeon, Korea). The thiol-modified oligonucleotide was de-protected before loading onto GNPs. Dried oligonucleotide (40.2 nmol) was dissolved in PBS (pH 7.5), 10 μL 1 N DTT was incorporated to it, and this mixture was incubated for 15 min at 25 °C. Unreacted DTT and thiol fragments were extracted thrice using 50 μL ethyl acetate. Then, the de-protected oligonucleotide was mixed with 1 mL of 0.2 nM GNPs or RVT@PVP-GNPs. The solution was then mixed gently for 16 h at 4 °C to allow further conjugation. Subsequently, 1 mL of 0.5 × Tris borate EDTA (TBE) buffer was incorporated to the solution and centrifuged (9000×g, 10 min, 4 °C) to remove unconjugated oligonucleotides in the supernatant. Apt-RVT@PVP-GNPs concentration was estimated at 320 nm using a UV/visible spectrophotometer (Ultrospec 2100 pro).
Animal studies
After 2 weeks of acclimatization, 5 × 106 PANC-1 cells in 100 μL of DMEM/Matrigel (1:1, v/v) were subcutaneously inoculated into the right flanks of the BALB/c nu/nu mice. The experimental groups (n = 5) were segregated randomly after the tumor volumes reached approximately 80 mm3. PBS (150 μL) comprising GNPs or RVT@PVP-GNPs was intravenously injected into the tail veins of the mice twice a week. Tumor volumes, body weight, and food intake were monitored for 33 days. Tumor volume (V) was estimated according to the following formula: V = (a × b2)/2, where a and b are the length and width of each tumor, respectively. After 4 weeks of experiments, the tumors and kidneys were removed and sectioned for histopathological analysis via hematoxylin and eosin (H&E) staining. Tumors obtained from mice were subsequently homogenized and lysed for protein extraction as described above. The tumors and kidneys of mice were fixed in 4% paraformaldehyde after removal from mice. Then, the organs were embedded in paraffin and cut into 5-μm-thick sections. Subsequently, they were placed on polylysine-coated slides, stained with H&E, and observed under a microscope (Axiolab A1, Carl Zeiss, Oberkochen, Germany) at 200× magnification (Karsh et al. 2020).
Statistical analysis
Statistical analysis was conducted using SPSS Statistics 25 (IBM, Armonk, NY, USA). Student’s t-test was conducted to measure the statistical significance of differences between two groups. To assess the significance among three or more groups, one-way analysis of variance was conducted. As necessary, data were marked with *p < 0.05, **p < 0.01, or ***p < 0.001, which were considered statistically significant (Younus et al. 2019).
Availability of data and materials
The data sets generated or analyzed during this study are available on reasonable request.
References
Ahn S, Truong VNP, Kim B, Yoo M, Lim Y, Cho SK et al (2022) Design, synthesis, and biological evaluation of chalcones for anticancer properties targeting glycogen synthase kinase 3 beta. Appl Biol Chem 65:1–4
Al-Musawi S, Albukhaty S, Al-Karagoly H, Sulaiman GM, Jabir MS, Naderi-Manesh H (2020) Dextran-coated superparamagnetic nanoparticles modified with folate for targeted drug delivery of camptothecin. Adv Nat Sci-Nanosci 11:045009
Al-Salman HNK, Ali ET, Jabir M, Sulaiman GM, Al-Jadaan SAS (2020) 2-Benzhydrylsulfinyl-N-hydroxyacetamide-Naextracted from fig as a novel cytotoxic and apoptosis inducer in SKOV-3 and AMJ-13 cell lines via P53 and caspase-8 pathway. Eur Food Res Technol 246:1591–1608
Amrutkar M, Gladhaug IP (2017) Pancreatic cancer chemoresistance to gemcitabine. Cancers 9:157
El Barky AR, Mohamed TM, Ali EM, (2020) Detoxifying and antioxidant effect of ellagic acid nano particles in rats intoxicated with sodium nitrites. Appl Biol Chem 63:1–6
Ben Tahar I, Fickers P, Dziedzic A, Ploch D, Skora B, Kus-Liskiewicz M (2019) Green pyomelanin-mediated synthesis of gold nanoparticles: modelling and design, physico-chemical and biological characteristics. Microb Cell Fact 18:210
Boatright KM, Salvesen GS (2003) Mechanisms of caspase activation. Curr Opin Cell Biol 15:725–731
Bonnefont-Rousselot D (2016) Resveratrol and cardiovascular diseases. Nutrients 8:250
Boulares AH, Yakovlev AG, Ivanova V, Stoica BA, Wang G, Iyer S et al (1999) Role of poly (ADP-ribose) polymerase (PARP) cleavage in apoptosis: caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J Biol Chem 274(33):22932–22940
Buso D, Post M, Cantalini C, Mulvaney P, Martucci A (2008) Gold nanoparticle-Doped TiO2 semiconductor thin films: gas sensing properties. Adv Funct Mater 18:3843–3849
Chithrani BD, Ghazani AA, Chan WCW (2006) Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett 6:662–668
Chowdhury R, Ilyas H, Ghosh A, Ali H, Ghorai A, Midya A et al (2017) Multivalent gold nanoparticle-peptide conjugates for targeting intracellular bacterial infections. Nanoscale 9:14074–14093
Condorelli F, Salomoni P, Cotteret S, Cesi V, Srinivasula SM, Alnemri ES et al (2001) Caspase cleavage enhances the apoptosis-inducing effects of BAD. Mol Cell Biol 21:3025–3036
Dive C, Gregory CD, Phipps DJ, Evans DL, Milner AE, Wyllie AH (1992) Analysis and discrimination of necrosis and apoptosis (programmed cell-death) by multiparameter flow-cytometry. Biochim Biophys Acta 1133:275–285
Elahi N, Kamali M, Baghersad MH (2018) Recent biomedical applications of gold nanoparticles: a review. Talanta 184:537–556
Gangwar RK, Dhumale VA, Kumari D, Nakate UT, Gosavi SW, Sharma RB et al (2012) Conjugation of curcumin with PVP capped gold nanoparticles for improving bioavailability. Mat Sci Eng C-Mater 32:2659–2663
Germain M, Caputo F, Metcalfe S, Tosi G, Spring K, Aslund AKO et al (2020) Delivering the power of nanomedicine to patients today. J Control Release 326:164–171
Ghosh P, Han G, De M, Kim CK, Rotello VM (2008) Gold nanoparticles in delivery applications. Adv Drug Deliver Rev 60:1307–1315
Gimenez-Bastida JA, Avila-Galvez MA, Espin JC, Gonzalez-Sarrias A (2019) Conjugated physiological resveratrol metabolites induce senescence in breast cancer cells: role of p53/p21 and p16/Rb pathways, and ABC transporters. Mol Nutr Food Res 63:1900629
Haupt S, Berger M, Goldberg Z, Haupt Y (2003) Apoptosis—the p53 network. J Cell Sci 116:4077–4085
He X, Andersson G, Lindgren U, Li Y (2010) Resveratrol prevents RANKL-induced osteoclast differentiation of murine osteoclast progenitor RAW 264.7 cells through inhibition of ROS production. Biochem Bioph Res Co 401:356–362
Hockel M, Vaupel P (2001) Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. Jnci-J Natl Cancer I 93:266–276
Ibrahim AA, Kareem MM, Al-Noor TH, Al-Muhimeed T, AlObaid AA, Albukhaty S et al (2021) Pt(II)-thiocarbohydrazone complex as cytotoxic agent and apoptosis inducer in Caov-3 and HT-29 cells through the P53 and Caspase-8 pathways. Pharm Base 14:509
Jabir MS, Nayef UM, Abdulkadhim WK, Taqi ZJ, Sulaiman GM, Sahib UI et al (2021) Fe3O4 nanoparticles capped with PEG induce apoptosis in breast cancer AMJ13 cells via mitochondrial damage and reduction of NF-kappa B translocation. J Inorg Organomet 31:1241–1259
Jannin B, Menzel M, Berlot JP, Delmas D, Lancon A, Latruffe N (2004) Transport of resveratrol, a cancer chemopreventive agent, to cellular targets: plasmatic protein binding and cell uptake. Biochem Pharmacol 68:1113–1118
Jihad MA, Noori FTM, Jabir MS, Albukhaty S, AlMalki FA, Alyamani AA (2021) Polyethylene glycol functionalized graphene oxide nanoparticles loaded with nigella sativa extract: a smart antibacterial therapeutic drug delivery system. Molecules 26:3067
Joe AK, Liu H, Suzui M, Vural ME, Xiao DH, Weinstein IB (2002) Resveratrol induces growth inhibition, S-phase arrest, apoptosis, and changes in biomarker expression in several human cancer cell lines. Clin Cancer Res 8:893–903
Kanduc D, Mittelman A, Serpico R, Sinigaglia E, Sinha AA, Natale C et al (2002) Cell death: apoptosis versus necrosis (review). Int J Oncol 21:165–170
Karsh EH, Kadhim RJ, and Jabir MS (2020) Effect of graphene oxide and gold nanoparticles on kidney parameters of male mice. In AIP Conference Proceedings 2213
Khashan KS, Abdulameer FA, Jabir MS, Hadi AA, Sulaiman GM (2020) Anticancer activity and toxicity of carbon nanoparticles produced by pulsed laser ablation of graphite in water. Adv Nat Sci-Nanosci 11:035010
Kim JS, Cuong DM, Bae YB, Cho SK (2022) Antioxidant and antiproliferative activities of solvent fractions of broccoli (Brassica oleracea L.) sprout. Appl Biol Chem 65:1
Kong FY, Zhang JW, Li RF, Wang ZX, Wang WJ, Wang W (2017) Unique roles of gold nanoparticles in drug delivery, targeting and imaging applications. Molecules 22:1445
Kotha A, Sekharam M, Cilenti L, Siddiquee K, Khaled A, Zervos AS et al (2006) Resveratrol inhibits Src and Stat3 signaling and induces the apoptosis of malignant cells containing activated Stat3 protein. Mol Cancer Ther 5:621–629
Lee DG, Go EB, Lee M, Pak PJ, Kim JS, Chung N (2019) Gold nanoparticles conjugated with resveratrol induce cell cycle arrest in MCF-7 cell lines. Appl Biol Chem 62:1
Li JL, Wang L, Liu XY, Zhang ZP, Guo HC, Liu WM et al (2009) In vitro cancer cell imaging and therapy using transferrin-conjugated gold nanoparticles. Cancer Lett 274:319–326
Li WP, Li CM, Ma LJ, Jin FG (2020) Resveratrol inhibits viability and induces apoptosis in the small-cell lung cancer H446 cell line via the PI3K/Akt/c-Myc pathway. Oncol Rep 44:1821–1830
Liang ZJ, Wan Y, Zhu DD, Wang MX, Jiang HM, Huang DL et al (2021) Resveratrol mediates the apoptosis of triple negative breast cancer cells by reducing POLD1 expression. Front Oncol 11:569295
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25:402–408
Ma DSL, Tan LTH, Chan KG, Yap WH, Pusparajah P, Chuah LH et al (2018) Resveratrol-potential antibacterial agent against foodborne pathogens. Front Pharmacol 9:102
Martin SJ (2016) Cell death and inflammation: the case for IL-1 family cytokines as the canonical DAMPs of the immune system. Febs J 283:2599–2615
Mizrahi JD, Surana R, Valle JW, Shroff RT (2020) Pancreatic cancer. Lancet 395:2008–2020
Nechushtan A, Smith CL, Lamensdorf I, Yoon SH, Youle RJ (2001) Bax and Bak coalesce into novel mitochondria-associated clusters during apoptosis. J Cell Biol 153:1265–1276
Pearce AK, O’Reilly RK (2019) Insights into Active targeting of nanoparticles in drug delivery: advances in clinical studies and design considerations for cancer nanomedicine. Bioconjugate Chem 30:2300–2311
Qiao WL, Wang BC, Wang YZ, Yang LC, Zhang YQ, and Shao PY (2010) Cancer therapy based on nanomaterials and nanocarrier systems. J Nanomater 2010
Rashid TM, Nayef UM, Jabir MS, Mutlak FAH (2021) Synthesis and characterization of Au:ZnO (core:shell) nanoparticles via laser ablation. Optik 244:167569
Roy SK, Chen QH, Fu JS, Shankar S, Srivastava RK (2011) Resveratrol Inhibits growth of orthotopic pancreatic tumors through activation of FOXO transcription factors. PLoS ONE 6:e25166
Sengupta S, Harris CC (2005) p53: Traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Bio 6:44–55
Shah NB, Dong JP, Bischof JC (2011) Cellular uptake and nanoscale localization of gold nanoparticles in cancer using label-free confocal Raman microscopy. Mol Pharmaceut 8:176–184
Shi J, Kantoff PW, Wooster R, Farokhzad OC (2017) Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer 17:20–37
Singh P, Pandit S, Mokkapati VRSS, Garg A, Ravikumar V, Mijakovic I (2018) Gold nanoparticles in diagnostics and therapeutics for human cancer. Int J Mol Sci 19:1979
Soundararajan S, Chen WW, Spicer EK, Courtenay-Luck N, Fernandes DJ (2008) The nucleolin targeting aptamer AS1411 destabilizes bcl-2 messenger RNA in human breast cancer cells. Can Res 68:2358–2365
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-Cancer J Clin 71:209–249
Turkevich J, Stevenson PC, Hillier J (1951) A study of the nucleation and growth processes in the synthesis of colloidal gold. Discuss Faraday Soc 55:55–75
Utaipan T, Boonyanuphong P, Chuprajob T, Suksamrarn A, Chunglok W (2020) A trienone analog of curcumin, 1,7-bis(3-hydroxyphenyl)-1,4,6-heptatrien-3-one, possesses ROS- and caspase-mediated apoptosis in human oral squamous cell carcinoma cells in vitro. Appl Biol Chem 63:1–11
Villa P, Kaufmann SH, Earnshaw WC (1997) Caspases and caspase inhibitors. Trends Biochem Sci 22:388–393
Vines JB, Yoon JH, Ryu NE, Lim DJ, Park H (2019) Gold nanoparticles for photothermal cancer therapy. Front Chem 7:167
Wang DD, Xu XW, Zhang K, Sun B, Wang L, Meng L et al (2018) Codelivery of doxorubicin and MDR1-siRNA by mesoporous silica nanoparticles-polymerpolyethylenimine to improve oral squamous carcinoma treatment. Int J Nanomed 13:187–198
Weyermann J, Lochmann D, Zimmer A (2005) A practical note on the use of cytotoxicity assays. Int J Pharmaceut 288:369–376
Wilson WR, Hay MP (2011) Targeting hypoxia in cancer therapy. Nat Rev Cancer 11:393–410
Wuithschick M, Birnbaum A, Witte S, Sztucki M, Vainio U, Pinna N et al (2015) Turkevich in new robes: key questions answered for the most common gold nanoparticle synthesis. ACS Nano 9:7052–7071
Yang K, Hitomi M, Stacey DW (2006) Variations in cyclin D1 levels through the cell cycle determine the proliferative fate of a cell. Cell Div 1:32
Yoo J, Park C, Yi G, Lee D, Koo H (2019) Active targeting strategies using biological ligands for nanoparticle drug delivery systems. Cancers 11:640
Younus A, Al-Ahmer S, Jabir M (2019) Evaluation of some immunological markers in children with bacterial meningitis caused by Streptococcus pneumoniae. Res J Biotech 14:131–133
Yuan HY, Li J, Bao G, Zhang SL (2010) Variable nanoparticle-cell adhesion strength regulates cellular uptake. Phys Rev Lett 105:138101
Zhang XD, Wu D, Shen X, Liu PX, Yang N, Zhao B et al (2011) Size-dependent in vivo toxicity of PEG-coated gold nanoparticles. Int J Nanomed 6:2071–2081
Zhang Y, Yang D, Yang B, Li BY, Guo J, Xiao CL (2019) PM2.5 induces cell cycle arrest through regulating mTOR/P70S6K1 signaling pathway. Exp Ther Med 17:4371–4378
Acknowledgements
Not applicable.
Funding
This study was supported by a grant from the National Research Foundation of Korea (Grant No. NRF-2015R1A2A2A0100650).
Author information
Authors and Affiliations
Contributions
DG conducted the experiments, analyzed the data, and wrote the paper. MD and EB conducted the experiments and interpreted the data. NC supervised the research and interpreted the data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All applicable international guidelines for the care and use of animals were followed. Research on experimental animals was carried out with the consent of the local ethics committee for animal experiments.
Consent to publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Supplementary analysis for LDH leakage level, quantitative immunoblotting of cyclin D1 and sequence of AS1411 aptamer.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lee, D.G., Lee, M., Go, E.B. et al. Resveratrol-loaded gold nanoparticles enhance caspase-mediated apoptosis in PANC-1 pancreatic cells via mitochondrial intrinsic apoptotic pathway. Cancer Nano 13, 34 (2022). https://doi.org/10.1186/s12645-022-00143-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12645-022-00143-w