Abstract
Purpose
Testicular germ cell tumours (TGCTs) is the most common malignancy among young adult males. The etiology is multifactorial and both environmental and genetic factors play an important role in the origin and development of TGCT. Genetic susceptibility may result from the interaction of multiple common and low-penetrance genetic variants and one of the main candidate genes is PDE11A. Many PDE11A polymorphisms were found responsible for a reduced PDE activity in TGCT patients, who often also display impaired hormone and sperm profile. The aim of this study was to investigate testicular function and PDE11A sequence in testicular cancer cases.
Methods
Semen analysis was performed in 116 patients with unilateral and bilateral sporadic TGCTs and in 120 cancer-free controls. We also investigated hormone profile and PDE11A polymorphisms using peripheral blood samples.
Results
Our data revealed that TGCT patients showed lower testosterone levels, higher gonadotropins levels and worse semen quality than controls, although the mean and the medians of sperm parameters are within the reference limits. PDE11A sequencing detected ten polymorphisms not yet associated with TGCTs before. Among these, G223A in homozygosity and A288G in heterozygosity were significantly associated with a lower risk of testicular tumour and they displayed a positive correlation with total sperm number.
Conclusions
Our findings highlight the key role of PDE11A in testis and suggest the presence of an underlying complex and fine molecular mechanism which controls testis-specific gene expression and susceptibility to testicular cancer.
Similar content being viewed by others
Introduction
Testicular germ cell tumours (TGCTs) represent the most common solid malignancy in men of reproductive age with an initial peak in childhood and a second, much larger peak beginning immediately after puberty [1]. They comprise about 95% of testicular tumours and their incidence has increased 3–4 times over the last 50 years. TGCTs are classified into two categories based on the presence of one or more histological types: tumours with a single histological type (seminomas and non-seminomas) that represent about 40% of all testicular neoplasms and tumours displaying two or more histological types (mixed tumours) that are the remaining 60%.
Histopathological studies have shown that most TGCTs arise from germ cell neoplasia in situ [2]. Dynamic epigenetic changes occur during normal development of germ cells and expression of genes which regulate this process is tightly controlled by epigenomic mechanisms, such as DNA methylation and microRNA [3,4,5,6,7]. Therefore, any genetic and environmental factor disturbing the maturation of primordial germ cells (PGCs) or gonocytes could induce the onset of TGCTs.
TGCTs can affect one (unilateral) or both testes (bilateral) simultaneously (synchronous forms) or after a certain period of time from the first testicular manifestation (metachronous forms) [8]. The presence of a unilateral testicular tumour is one of the most important risk factor for the development of a malignant cancer in the contralateral organ [9]. As bilateral testicular tumours are often diagnosed in young men with a familiarity for TGCTs, a genetic component can be present at the base of these neoplasms. However, the etiology is multifactorial and, in addition to the genetic contribution, risk factors, such as exposure to endocrine disruptors (EDs) and cryptorchidism, could play a key role in carcinogenesis. In particular, EDs could cause the development of TGCTs by disturbing the synthesis, release, transport, metabolism, binding, action or elimination of endogenous hormones during embryonic development [10, 11].
Although the molecular causes of these tumours remain elusive, evidences in literature support the hypothesis of a genetic contribution for the development of TGCTs with PDE11A as a possible candidate gene [12,13,14]. This gene encodes a dual-specificity phosphodiesterase (PDE) capable of hydrolyzing both cAMP and cGMP and isoform 4 is highly expressed in testicular tissue, which so far is the only known tissue expressing all four PDE11A isoforms [15]. Pde11a knockout mice display male infertility, a factor associated to increased TGCT risk [16,17,18,19]. Moreover, alterations in the cAMP pathway have also been observed in non-germ cell-derived testicular tumours [20, 21], suggesting the importance of this pathway in testicular tissue. Interestingly, two recent studies investigated the putative link between testicular tumours and PDE11A polymorphisms (SNPs), finding that inactivating PDE11A variants seem to be associated with TGCT risk in both familial and sporadic cases [12, 14].
In the light of the aforementioned evidences, the aim of our study was:
-
a.
to evaluate hormone profile and sperm parameters to assess testicular function in 116 patients with unilateral and bilateral sporadic TGCTs compared to 120 cancer-free controls;
-
b.
to investigate PDE11A SNPs in our caseload to confirm the association between testicular tumours and two missense variants identified in previous studies, p.V820M and p.K568R.
Materials and methods
Patients
The study was approved by our University Hospital’s Institutional Review Board (Ethical Committee of “Sapienza” University of Rome—Azienda Ospedaliera Policlinico Umberto I) and all patients gave their informed written consent.
We enrolled 116 Caucasian patients with unilateral or bilateral sporadic TGCTs (Group T) attending the Laboratory of Seminology—Sperm Bank “Loredana Gandini”, Department of Experimental Medicine at “Sapienza” University of Rome for semen cryopreservation. As controls, we recruited 120 cancer-free Caucasian men (Group C), attending our laboratory for semen analysis as part of an andrological work-up for preconceptional screening. Men with a history of azoospermia, hypogonadism and known genetic diseases were excluded from the study. For the two groups, we estimated both sperm parameters and serum hormone levels. Moreover, we performed molecular analysis to identify PDE11A SNPs.
Semen analysis
Semen samples were collected by masturbation after 3–5 days of abstinence. All samples were allowed to liquefy at 37 °C for 60 min and were then assessed according to WHO 2010 [22]. The following variables were taken into consideration: ejaculate volume (ml), sperm concentration (106 per ml), total sperm number (106 per ejaculate), progressive motility (%) and morphology (% abnormal forms).
Hormone profile
A peripheral blood sample was collected from each subject at 8 a.m. after overnight fasting to measure serum levels of follicle-stimulating hormone (FSH), luteinizing hormone (LH) and testosterone. Hormones were quantified by Chemiluminescent Microparticle ImmunoAssay (CMIA, Architect System; Abbott Laboratories, Abbott Park, IL, USA), with detection limits of 0.05 UI/L, 0.07 UI/L, 0.28 nmol/L for FSH, LH and testosterone, respectively. Intra and inter-assay coefficients of variation were 3.1% and 7.0% at 3.2 UI/L (FSH), 3.6% and 5.1% at 3.3 UI/L (LH), 2.1% and 3.6% at 10.08 nmol/L (testosterone), respectively. In our laboratory, normal ranges for adults were 1.38–9.58 UI/L (FSH), 1.80–8.16 UI/L (LH) and 9.4–33.5 nmol/L (testosterone), respectively.
PDE11A sequencing
Blood samples underwent following steps to identify PDE11A SNPs. Firstly, DNA was extracted from peripheral blood leukocytes using Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). Extracted DNA was quantified by NanoDrop ND-2000 (Thermo Fisher Scientific, Waltham, MA, USA) and underwent molecular analysis to perform PDE11A sequencing based on Sanger method.
We identified a region of PDE11A in which the SNPs rs140269105 (p.V820M) and rs148955609 (p.K568R), previously identified in literature [12, 14], are present. Primer pairs were designed about 200 bp upstream and downstream of the 2 SNPs of interest using the software primer 3 plus. We refer to Fragment1 for the region containing the SNP p.V820M and Fragment2 for the region containing the SNP p.K568R. The primer sequences were as follows:
-
Fragment1_F 5′-GGGCTGTGCAATAAACTGTG-3’
-
Fragment1_R 5′-ATAAACAGTGCTGCCCCTTG-3’
-
Fragment2_F 5′-GAATGGGCTTCAAGGCATCT-3’
-
Fragment2_R 5′-ATGTGCCTATTTCCCCAAGG-3’
The amplification reaction was carried out using 30 ng of genomic DNA in 50 µl under the following PCR conditions: 10 min at 95 °C followed by 35 cycles of 1 min at 94 °C, 1 min at 58 °C, 1 min and 20 s at 72 °C and a final extension step at 72 °C for 12 min. A 5 µl of each PCR product was then used for electrophoresis on 2% agarose gel to check the presence and exact length of the amplified fragments (389 and 444 nucleotides for Fragment1 and Fragment2, respectively).
The fragments were then purified using PureLink PCR Purification Kit (Invitrogen, Life Tecnologies, USA) and the sequencing was performed with Big Dye Terminator® v1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). To separate the end-labeling reaction products from the unused dye-terminators, salts and other low molecular weight products, Centri-Sep® gel filtration columns (Applied Biosystems, USA) were used. Finally, the labelled fragments underwent capillary electrophoresis using the 3500 Genetic Analyzer sequencer (Applied Biosystems).
Bioinformatic analysis
Raw data from the capillary electrophoresis were analysed by Sequencing Analysis v5.1 (Applied Biosystem, USA). To improve reliability and exclude the presence of artefacts, each sample was sequenced twice using the same primers in the two directions 5′–3′ and 3′–5′. To detect PDE11A SNPs, samples sequences were compared against the reference sequence GRCh38.p12 (NG_012168.2) in GenBank using sequence alignment (Blast). All SNPs discovered were compared with the lists in dbSNP database (https://www.ncbi.nlm.nih.gov/snp). The impact of all SNPs detected on coding sequence was studied to understand if they were synonymous or not. Furthermore, we verified if they were already identified in the previous literature and if they were associated with pathological conditions. The software used was Geneious v.R 9.0.2.
Statistical analysis
Continuous variables are presented as mean ± SD or as median and interquartile range, as appropriate, after evaluation of the normality of distributions using the Kolmogorov–Smirnov test. Student’s T or Mann–Whitney U test have been used to compare sperm parameters between the two groups. Categorical variables are presented as counts and/or percentages and differences in frequencies are performed by the χ2 test. The presence of statistically significant correlations among sperm parameters and hormone levels was evaluated using Spearman’s rank correlation test.
Binary logistic regression models have been used to analyze associations between testicular cancer and PDE11A SNPs. Finally, associations between sperm parameters, hormone levels and polymorphisms have been investigated by univariate generalized linear models.
PDE11A alleles frequencies were determined by the gene counting method and the agreement of the genotype distribution with the Hardy–Weinberg Equilibrium test was calculated. Differences of the SNPs frequencies between the study groups have been evaluated by the χ2 test. A two-tailed P value ≤ 0.05 was considered significant. All computations were carried out with Statistical Package for the Social Sciences (SPSS) 25.0 (SPSS Inc., Chicago, USA).
Results
Study population and histological data
We studied 116 patients affected by unilateral or bilateral sporadic TGCTs (Group T) and 120 cancer-free controls (Group C), aged 32.6 ± 7.3 and 27.8 ± 7.2 years, respectively (P < 0.001). No significant differences between Group T and Group C were found in Body Mass Index (BMI) (24.7 ± 3.8 vs. 24.2 ± 3.2 kg/m2 respectively, P = 0.355) and percentage of smokers (25.0% vs. 21.7%, respectively, P = 0.647).
Group T was composed by:
-
5 patients with synchronous bilateral neoplasm;
-
13 patients with contralateral metachronous recurrence neoplasm;
-
98 patients with unilateral neoplasm (of which 65% was seminomas and the remaining 35% was mixed tumours and non-seminomas).
In particular, metachronous neoplasms appeared after a median of 4 years and these patients had been treated with 2–4 cycles of cisplatin, bleomycin and etoposide (PEB) in three cases, a single cycle of radiotherapy in other three cases and only follow up in the seven remaining cases.
Semen analysis
Comparison of the sperm parameters of the two study groups revealed a poor semen quality in Group T (Table 1). Except for the ejaculate volume (3.1 ± 1.6 vs. 2.9 ± 1.1 ml in Group T and Group C, respectively, P = 0.699), Group T showed a significantly lower total sperm number (93.5 ± 99.2 vs. 178.0 ± 171.5 × 106/ejaculate in Group T and Group C, respectively, P < 0.001), progressive motility (34.1 ± 19.3% vs. 42.3 ± 18.1%, in Group T and Group C, respectively, P < 0.001) and a higher percentage of abnormal forms (89.6 ± 13.3% vs. 87.7 ± 7.2% in Group T and Group C, respectively, P < 0.001). In particular, in Group T 40% of samples appears oligozoospermic in contrast to 23.5% of samples in Group C.
It is worth stressing that in both groups the means and the medians of sperm parameters are within the reference limits according to WHO 2010.
Furthermore, in the caseload as a whole, progressive motility and abnormal forms are weakly correlated with age (progressive motility: ρ = − 0.184, P = 0.004; abnormal forms: ρ = 0.220, P = 0.001) and, except for the morphology, with BMI (total sperm number: ρ = −0.153, P = 0.026; progressive motility: ρ = −0.144, P = 0.036).
Hormone parameters
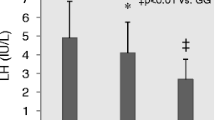
The hormone dosage revealed a significant worse profile in Group T (Table 2). In particular, testosterone level was significantly lower (17.7 ± 6.4 vs. 21.6 ± 6.5 nmol/ml in Group T and Group C, respectively, P < 0.001), while the levels of gonadotropins were significantly higher (FSH: 10.4 ± 7.2 vs. 4.1 ± 3.6 mUI/ml, P < 0.001; LH: 5.5 ± 4.2 vs. 3.8 ± 1.7 mUI/ml, P < 0.001, in Group T and Group C, respectively).
As expected, hormone levels were significantly correlated with both age (FSH: ρ = 0.439, P < 0.001; LH: ρ = 0.240, P = 0.001; testosterone: ρ = − 0.186, P = 0.010) and BMI (FSH: ρ = 0.180, P = 0.019; LH: ρ = 0.245, P = 0.001; testosterone: ρ = −0.260, P = 0.001).
Genetic study of PDE11A
PDE11A sequencing did not reveal the two missense variants investigated (p.V820M and p.K568R) in the caseload. However, we detected the following ten polymorphisms:
-
For the Fragment1: C207T, G223A, A288G, T366C;
-
For the Fragment2: C102A, G172A, C189T, T245C, C255A, G371C.
Sequencing analysis of Fragment1 was carried out in 116 patients with TGCTs and in 120 subjects from Group C. Instead, sequencing analysis of Fragment2 was performed in 99 patients affected by TGCTs and in 100 cancer-free controls.
Alleles frequencies were determined by the gene counting method, as reported in Tables 3 and 4. All the alleles resulted in accordance with the Hardy–Weinberg equilibrium.
Analysis of PDE11A SNPs revealed that all subjects (patients and controls) were homozygous for the SNPs C207T, T366C, G172A, T245C and C255A. Furthermore, the SNPs G223A and A288G appeared differently in cases and controls (χ2 P = 0.004 and P = 0.024, respectively). In contrast, the SNPs C102A, C189T and G371C showed a similar distribution in all subjects investigated.
Then we used binary logistic regression models to analyze associations between testicular cancer and polymorphisms, considering the SNPs G223A, A288G, C102A, C189T and G371C as independent variables. This analysis revealed that only the SNP G223A in homozygosity and A288G in heterozygosity were significantly associated with a lower risk of testicular tumour (G223A homozygous: OR 0.123, 95% CI 0.034–0.451, P = 0.002; A288G heterozygous: OR 0.199, 95% CI 0.068–0.578, P = 0.003), as reported in Fig. 1a, b.
Finally, using univariate models and after correction for FSH values, we identified an association between total sperm number and the SNPs G223A (Fig. 2a) and A288G (Fig. 2b) (P = 0.005 and P = 0.003, respectively; R2 = 0.241). In particular, total sperm number appeared higher in homozygote AA, in the case of G223A, and in heterozygote AG, in the case of A288G, than other genotypes.
Discussion
TGCT is the most common malignancy in young adult males and its incidence has increased over the last 50 years. The risk for the development of these neoplasms is higher in Scandinavia, Switzerland and Germany, intermediate in the United States, Britain and Mexico, lower in Africa and Asia [23, 24]. However, the reasons for the differences in the incidence of TGCT among different ethnic groups are unknown [25].
Clinical and epidemiological studies suggest that several etiological factors could increase the susceptibility to testicular cancer, such as a family history of testicular cancer, exposure to environmental pollutants (endocrine disruptors) and cryptorchidism [10, 11, 26,27,28,29,30]. As the underlying molecular causes still remain unclear, we aimed to investigate in TGCT patients the genetic contribution of PDE11A polymorphisms, one of the putative genes behind of this complex and multifactorial disease. Moreover, we evaluated sperm parameters and hormone profile to assess testicular function in TGCT patients compared to cancer-free controls.
Sperm parameters
Despite the mean sperm parameters of Group T are above the 5th percentile of the WHO reference value, comparison with the healthy controls revealed a poorer semen quality in our cohort of testicular cancer patients. As TGCTs seem to arise from germ cell neoplasia in situ, which could originate from PGCs or gonocytes whose maturation is disturbed [2], testicular neoplasms may induce male infertility as a consequence of sperm parameters alterations.
The poorer semen quality observed in our TGCT patients is consistent with literature evidence, although the relationship between testicular neoplasms and infertility appears complex and controversial due to genetic, environmental and ethnic differences which could impact spermatogenesis. Over the last 20 years several studies evaluated sperm characteristics in these neoplasms before treatment, showing an impaired semen quality in TGCT patients [31,32,33,34,35,36].
It is noteworthy that, except for a few studies [37, 38], semen quality appears more compromised in testicular neoplasms than in other malignancies, even before beginning any antineoplastic treatment [32,33,34, 36]. This could be caused by TGCT itself through hormonal alterations and metabolic settings. In particular, β-human chorionic gonadotropin (β-hCG) could influence spermatogenesis directly or indirectly through hypothalamus-pituitary-gonad axis. It has been speculated that β-hCG could exert LH-like effects on Leydig cells and could induce a feedback on hypothalamus-pituitary axis impairing gonadal function [32, 39]. As reported in the literature, the presence of a compromised spermatogenesis in TGCT patients with higher serum β-hCG levels would confirm this hypothesis [32, 39]. Malignancy might also result in malnutrition, with consequent psychological complications and deficiencies in vitamins and minerals needed for a proper testicular function. Finally, spermatogenesis might be negatively influenced by periods of fever and by tumour release of cytokines. All these factors expose testicular cancer patients at the highest risk of having reduced semen quality before treatment, which can further negatively impact fertility making sperm cryopreservation an important clinical option for male fertility preservation [32, 35, 36, 40,41,42,43,44,45,46,47,48,49,50,51,52,53].
Hormone profile
The presence of a tumour is supposedly associated with an altered hormone profile due to different causes, such as a dysregulated hormonal secretion or a release of hormones by the tumour itself.
In support of this hypothesis, patients affected by TGCTs are more likely to show higher levels of FSH and LH and lower levels of testosterone, an endocrine pattern which also characterizes infertile men [54]. It should be stressed that orchiectomy, testicular dysgenesis syndrome, treatment after orchiectomy and aging could play a key role in the increase of prevalence of hypogonadism in these patients.
The hormone profile appeared significantly altered in Group T in comparison with the cancer-free controls: in particular, testicular cancer patients showed higher serum gonadotropins levels with reduced testosterone, confirming previous literature observations [55, 56].
Although the hormone profile was altered, our cohort of testicular cancer patients showed mean sperm parameters lower than the controls but within the reference limits. As also demonstrated in animal models, this evidence can be explained through a compensatory mechanism: orchiectomy may cause a rapid decline in inhibin B levels due to the halving of the number of Sertoli cells; this provides the stimulus for a surge in FSH secretion by the pituitary which may induce proliferation of germ cells in contralateral testis and an increase of testicular volume, under physiological functional conditions [57,58,59]. For this reason, despite the known association between BMI and hypogonadotropic hypogonadism, we found that in this cohort of TGCT patients BMI is positively associated with gonadotropins.
PDE11A analysis
The frequent diagnosis in young men with a positive family history for TGCTs and the increased risk for the children and siblings of men with testicular cancer point to a genetic basis of these neoplasms [26,27,28].
Linkage analyses suggest that the susceptibility may result from the interaction of multiple common and low-penetrance genetic variants [60,61,62] and one of the main candidate genes is PDE11A, expressed in testicular tissue in all four known isoforms [15].
Studies of adrenal, prostate and testicular cancer have suggested that PDE11A variants may represent susceptibility modifiers rather than direct and sufficient causes of these neoplasms [63]. This gene may play a key role also in spermatogenesis and fertilization potential, as suggested by observation that Pde11a knockout mice displayed reduced sperm concentration, rate of forward progression, percentage of live spermatozoa and increased premature/spontaneous capacitance [17]. These evidences suggest a role for PDE11 in testicular tissue.
Inactivating PDE11A variants induce alterations in cAMP pathway increasing the levels of this cyclic nucleotide, which may promote TGCT development similarly to what has been observed in non-germ cell-derived testicular tumours, such as in Leydig cell hyperplasia, McCune-Albright syndrome and Carney complex-associated Sertoli cell tumours [20, 21].
The role of PDE11A polymorphisms has been explored in various diseases but recent studies highlighted their contribution also in testicular cancer. In 2009, Horvath et al. analyzed the PDE11A coding sequence in 95 patients with familial and bilateral TGCT, finding a significantly higher frequency of the non-synonymous substitution p.V820M among testicular cancer patients than control subjects [12].
Subsequently, Pathak et al. sequenced the PDE11A coding region in 259 patients with both familial and sporadic TGCT, detecting 55 variants including p.V820M and p.K568R, which were present only in cases and not in controls [14]. It is noteworthy that PDE11A variants identified in these studies resulted in reduced PDE activity and increased cAMP levels modifying the TGCT risk not only in familial and bilateral form, but also in sporadic form.
In our caseload, we aimed to identify p.V820M and p.K568R, two polymorphisms detected in the aforementioned studies, to confirm their role in patients affected by unilateral and bilateral sporadic TGCTs. Both SNPs affect critical sites of the enzyme: in particular, p.V820M (Fragment1) is placed in the catalytic domain, while p.K568R (Fragment2) in GAF-B domain required for enzyme oligomerization.
None of our TGCT patients and controls showed the two SNPs investigated. However, PDE11A sequencing revealed ten new polymorphisms not yet associated with testicular cancer before: four for the Fragment1 (C207T, G223A, A288G, T366C) and six for the Fragment2 (C102A, G172A, C189T, T245C, C255A, G371C).
The discrepancies in the genetic results between our study and literature could arise from differences in the alleles frequencies due to geographical distribution. Although the populations in question have Caucasian origin, environmental factors and genetic recombination may have diversified the genetic profiles over time. Furthermore, it should be stressed that we analyzed almost exclusively patients affected by unilateral sporadic TGCTs, whereas the studies reported in the literature focused mainly on bilateral familial cases.
As most of the new SNPs detected in our study are uniformly present in the caseload as a whole, it is plausible that they are constitutive polymorphisms. The only two SNPs showing a different significant distribution between case and controls are G223A and A288G, both localized in the Fragment1 such as p.V820M detected in the aforesaid studies. In particular, A288G is an intronic variant, while G223A is not present in dbSNP database. Therefore, it was not possible to identify it as an intronic or exonic variant. Examining putative associations between these two SNPs and pathological conditions, we found that only A288G has previously been related to antidepressant treatment response [64].
Analysis of associations between testicular cancer and PDE11A polymorphisms revealed that the homozygote AA, in the case of G223A, and the heterozygote AG, in the case of A288G, were significantly associated with a lower risk of testicular tumour than the other genotypes. Moreover, they displayed a significant positive correlation with total sperm number. As these two genotypes resulted associated with a lower risk of TGCTs, we suggest that they could improve PDE11A function in the presence of risk factors for testicular cancer development, such as cryptorchidism, endocrine disruptors, etc. Hence, this function would be opposite to that induced by the SNPs detected by Horvath et al. 2009 [12] and Pathak et al. 2015 [14], which reduce enzymatic activity increasing cAMP levels and TGCT risk. The putative protective role of these two genotypes can be deduced from the finding of reduced PDE activity and consequent increased cAMP levels which also characterize other tumour settings [20, 21].
Moreover, the association with total sperm number allows us to hypothesize that these genetic variants could influence, not only the onset of testicular neoplasms, but also the spermatogenesis process.
However, as the underlying molecular mechanisms are still unclear, it is plausible to assume that additional factors involved in cAMP signaling could play a pivotal role. An example is provided by CREM (cAMP-response-element modulator), a transcription factor responsive to the cAMP signal transduction pathway which represents a master regulator of key testis-specific genes necessary for spermatogenesis [65, 66].
Conclusions
TGCTs are complex neoplasms whose aetiology is multifactorial. In our study, we observed that TGCT patients showed an altered hormone profile and a poor semen quality, although the sperm parameters were within the reference limits. Moreover, we identified ten new PDE11A polymorphisms, two of which significantly associated with a lower risk of testicular tumour. This result remarks that the genetic contribution could be critical in the susceptibility to these neoplasms.
Nonetheless, PDE11A role in testis is still unclear and the contribution of additional factors involved in cAMP signaling not investigated in our study cannot be excluded. Hence, further investigations are needed to elucidate the underlying molecular mechanisms and to clarify how alterations in cAMP pathway could influence the TGCT risk.
References
Bosetti C, Bertuccio P, Chatenoud L, Negri E, La Vecchia C, Levi F (2011) Trends in mortality from urologic cancers in Europe, 1970–2008. Eur Urol 60(1):1–15
Fukawa T, Kanayama HO (2018) Current knowledge of risk factors for testicular germ cell tumors. Int J Urol 25(4):337–344
Messerschmidt DM, Knowles BB, Solter D (2014) DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev 28(8):812–828
Murray MJ, Nicholson JC, Coleman N (2015) Biology of childhood germ cell tumours, focussing on the significance of microRNAs. Andrology 3(1):129–139
Murray MJ, Huddart RA, Coleman N (2016) The present and future of serum diagnostic tests for testicular germ cell tumours. Nat Rev Urol 13(12):715–725
Fendler A, Stephan C, Yousef GM, Kristiansen G, Jung K (2016) The translational potential of microRNAs as biofluid markers of urological tumours. Nat Rev Urol 13(12):734–752
Pelloni M, Coltrinari G, Paoli D, Pallotti F, Lombardo F, Lenzi A, Gandini L (2017) Differential expression of miRNAs in the seminal plasma and serum of testicular cancer patients. Endocrine 57(3):518–527
Al-Obaidy KI, Trevino KE, Idrees MT (2019) Clinicopathologic characterization of bilateral testicular germ cell tumors with immunohistochemical evaluation of mismatch repair and BRAF (V600E) genes mutations. Int J Surg Pathol 27(6):619–623
Sharma P, Dhillon J, Sexton WJ (2015) Intratubular germ cell neoplasia of the testis, bilateral testicular cancer, and aberrant histologies. Urol Clin North Am 42(3):277–285
Landero-Huerta DA, Vigueras-Villasenor RM, Yokoyama-Rebollar E, Arechaga-Ocampo E, Rojas-Castaneda JC, Jimenez-Trejo F, Chavez-Saldana M (2017) Epigenetic and risk factors of testicular germ cell tumors: a brief review. Front Biosci (Landmark Ed) 22:1073–1098
Pallotti F, Pelloni M, Gianfrilli D, Lenzi A, Lombardo F, Paoli D (2020) Mechanisms of testicular disruption from exposure to bisphenol a and phtalates. J Clin Med 9(2):471
Horvath A, Korde L, Greene MH, Libe R, Osorio P, Faucz FR, Raffin-Sanson ML, Tsang KM, Drori-Herishanu L, Patronas Y, Remmers EF, Nikita ME, Moran J, Greene J, Nesterova M, Merino M, Bertherat J, Stratakis CA (2009) Functional phosphodiesterase 11A mutations may modify the risk of familial and bilateral testicular germ cell tumors. Cancer Res 69(13):5301–5306
Greene MH, Kratz CP, Mai PL, Mueller C, Peters JA, Bratslavsky G, Ling A, Choyke PM, Premkumar A, Bracci J, Watkins RJ, McMaster ML, Korde LA (2010) Familial testicular germ cell tumors in adults: 2010 summary of genetic risk factors and clinical phenotype. Endocr Relat Cancer 17(2):R109-121
Pathak A, Stewart DR, Faucz FR, Xekouki P, Bass S, Vogt A, Zhang X, Boland J, Yeager M, Loud JT, Nathanson KL, McGlynn KA, Stratakis CA, Greene MH, Mirabello L (2015) Rare inactivating PDE11A variants associated with testicular germ cell tumors. Endocr Relat Cancer 22(6):909–917
Horvath A, Boikos S, Giatzakis C, Robinson-White A, Groussin L, Griffin KJ, Stein E, Levine E, Delimpasi G, Hsiao HP, Keil M, Heyerdahl S, Matyakhina L, Libè R, Fratticci A, Kirschner LS, Cramer K, Gaillard RC, Bertagna X, Carney JA, Bertherat J, Bossis I, Stratakis CA (2006) A genome-wide scan identifies mutations in the gene encoding phosphodiesterase 11A4 (PDE11A) in individuals with adrenocortical hyperplasia. Nat Genet 38(7):794–800
Jacobsen R, Bostofte E, Engholm G, Hansen J, Olsen JH, Skakkebaek NE, Moller H (2000) Risk of testicular cancer in men with abnormal semen characteristics: cohort study. BMJ 321(7264):789–792
Wayman C, Phillips S, Lunny C, Webb T, Fawcett L, Baxendale R, Burgess G (2005) Phosphodiesterase 11 (PDE11) regulation of spermatozoa physiology. Int J Impot Res 17(3):216–223
Negri L, Benaglia R, Fiamengo B, Pizzocaro A, Albani E, Levi Setti PE (2008) Cancer risk in male factor-infertility. Placenta 29(Suppl B):178–183
Paoli D, Gilio B, Piroli E, Gallo M, Lombardo F, Dondero F, Lenzi A, Gandini L (2009) Testicular tumors as a possible cause of antisperm autoimmune response. Fertil Steril 91(2):414–419
Weinstein LS, Shenker A, Gejman PV, MerinoMJ FE, Spiegel AM (1991) Activating mutations of the stimulatory G protein in the McCune–Albright syndrome. N Engl J Med 325(24):1688–1695
Kirschner LS, Carney JA, Pack SD, Taymans SE, Giatzakis C, Cho YS, Cho-Chung YS, Stratakis CA (2000) Mutations of the gene encoding the protein kinase A type I-alpha regulatory subunit in patients with the Carney complex. Nat Genet 26(1):89–92
WHO (2010) WHO laboratory manual for the examination and processing of human semen, 5th edn. World Health Organization, Geneva
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61(2):69–90
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Bray F, Ferlay J, Devesa SS, McGlynn KA, Moller H (2006) Interpreting the international trends in testicular seminoma and nonseminoma incidence. Nat Clin Pract Urol 3(10):532–543
Dieckmann KP, Pichlmeier U (1997) The prevalence of familial testicular cancer: an analysis of two patient populations and a review of the literature. Cancer 80(10):1954–1960
Swerdlow AJ, De Stavola BL, Swanwick MA, Maconochie NE (1997) Risks of breast and testicular cancers in young adult twins in England and Wales: evidence on prenatal and genetic aetiology. Lancet 350(9093):1723–1728
Hemminki K, Li X (2004) Familial risk in testicular cancer as a clue to a heritable and environmental aetiology. Br J Cancer 90(9):1765–1770
Markey CM, Rubin BS, Soto AM, Sonnenschein C (2002) Endocrine disruptors: from Wingspread to environmental developmental biology. J Steroid Biochem Mol Biol 83(1–5):235–244
Paoli D, Giannandrea F, Gallo M, Turci R, Cattaruzza MS, Lombardo F, Lenzi A, Gandini L (2015) Exposure to polychlorinated biphenyls and hexachlorobenzene, semen quality and testicular cancer risk. J Endocrinol Invest 38(7):745–752
Hallak J, Kolettis PN, Sekhon VS, Thomas AJ Jr, Agarwal A (1999) Sperm cryopreservation in patients with testicular cancer. Urology 54(5):894–899
Petersen PM, Skakkebaek NE, Vistisen K, Rørth M, Giwercman A (1999) Semen quality and reproductive hormones before orchiectomy in men with testicular cancer. J Clin Oncol 17(3):941–947
Gandini L, Lombardo F, Salacone P, Paoli D, Anselmo AP, Culasso F, Dondero F, Lenzi A (2003) Testicular cancer and Hodgkin’s disease: evaluation of semen quality. Hum Reprod 18(4):796–801
Bizet P, Saias-Magnan J, Jouve E, Grillo JM, Karsenty G, Metzler-Guillemain C, Perrin J (2012) Sperm cryopreservation before cancer treatment: a 15-year monocentric experience. Reprod Biomed Online 24(3):321–330
Bujan L, Walschaerts M, Moinard N, Hennebicq S, Saias J, Brugnon F, Auger J, Berthaut I, Szerman E, Daudin M, Rives N (2013) Impact of chemotherapy and radiotherapy for testicular germ cell tumors on spermatogenesis and sperm DNA: a multicenter prospective study from the CECOS network. Fertil Steril 100(3):673–680
Caponecchia L, Cimino G, Sacchetto R, Fiori C, Sebastianelli A, Salacone P, Marcucci I, Tomassini S, Ragoet R (2015) Do malignant diseases affect semen quality? Sperm parameters of men with cancers. Andrologia 48(3):333–340
Chung K, Irani J, Knee G, Efymow B, Blasco L, Patrizio P (2004) Sperm cryopreservation for male patients with cancer: an epidemiological analysis at the University of Pennsylvania. Eur J of Obstet Gynecol Reprod Biol 113(Suppl 1):S7-11
Meseguer M, Molina N, García-Velasco JA, Remohí J, Pellicer A, Garrido N (2006) Sperm cryopreservation in oncological patients: a 14-year follow-up study. Fertil Steril 85(3):640–645
de Bruin D, de Jong IJ, Arts EG, Dullaart RPF, Sluiter WJ, Hoekstra HJ, Sleijfer DT, Gietema JA (2009) Semen quality in men with disseminated testicular cancer: relation with human chorionic gonadotropin beta-subunit and pituitary gonadal hormones. Fertil Steril 91(6):2481–2486
Hendry WF, Stedronska J, Jones CR, Blackmore CA, Barrett A, Peckham MJ (1983) Semen analysis in testicular cancer and Hodgkin’s disease: pre- and post-treatment findings and implications for cryopreservation. Br J Urol 55(6):769–773
Fritz K, Weissbach L (1985) Sperm parameters and ejaculation before and after operative treatment of patients with germ-cell testicular cancer. Fertil Steril 43(3):451–454
Fossa SD, Aass N, Molne K (1989) Is routine pre-treatment cryopreservation of semen worthwhile in the management of patients with testicular cancer? Br J Urol 64(5):524–529
Agarwal A, Tolentino MV Jr, Sidhu RS, Ayzman I, Lee JC, Thomas AJ Jr, Shekarriz M (1995) Effect of cryopreservation on semen quality in patients with testicular cancer. Urology 46(3):382–389
Meirow D, Schenker JG (1995) Cancer and male infertility. Hum Reprod 10(8):2017–2022
Kliesch S, Bergmann M, Hertle L, Nieschlag E, Behre HM (1997) Semen parameters and testicular pathology in men with testicular cancer and contralateral carcinoma in situ or bilateral testicular malignancies. Hum Reprod 12(12):2830–2835
Botchan A, Hauser R, Yogev L, Gamzu R, Paz G, Lessino JB, Yavetz H (1997) Testicular cancer and spermatogenesis. Hum Reprod 12(4):755–758
Lass A, Akagbosu F, Abusheikha N, Hassouneh M, Blayney M, Avery S, Brinsden P (1998) A programme of semen cryopreservation for patients with malignant disease in a tertiary infertility centre: lessons from 8 years’ experience. Hum Reprod 13(11):3256–3261
Panidis D, Rousso D, Stergiopoulos K, Papathanasiou K, Delkos D, Papaletsos M (1999) The effect of testicular seminoma in semen quality. Eur J of Obstet Gynecol Reprod Biol 83(2):219–222
Fitoussi O, Eghbali H, Tchen N, Berjon JP, Soubeyran P, Hoerni B (2000) Semen analysis and cryopreservation before treatment in Hodgkin’s disease. Ann Oncol 11(6):679–684
van Casteren NJ, Boellaard WP, Romijn JC, Dohle GR (2009) Gonadal dysfunction in male cancer patients before cytotoxic treatment. Int J Androl 33(1):73–79
Dohle GR (2010) Male infertility in cancer patients: review of the literature. Int J Urol 17(4):327–331
Trost LW, Brannigan RE (2012) Oncofertility and the male cancer patient. Curr Treat Options Oncol 13(2):146–160
Ostrowski KA, Walsh TJ (2015) Infertility with testicular cancer. Urol Clin North Am 42(3):409–420
Sineath RC, Mehta A (2019) Preservation of Fertility in Testis Cancer Management. Urol Clin North Am 46(3):341–351
Sprauten M, Brydøy M, Haugnes HS, Cvancarova M, Bjøro T, Bjerner J, Fosså SD, Oldenburg J (2014) Longitudinal serum testosterone, luteinizing hormone, and follicle-stimulating hormone levels in a population-based sample of long-term testicular cancer survivors. J Clin Oncol 32(6):571–578
Petrozzi A, Pallotti F, Pelloni M, Anzuini A, Radicioni AF, Lenzi A, Paoli D, Lombardo F (2019) Inhibin B: are modified ranges needed for orchiectomised testicular cancer patients? Asian J Androl 21(4):332–336
Medhamurthy R, Aravindan GR, Moudgal NR (1993) Hemiorchidectomy leads to dramatic and immediate alterations in pituitary follicle stimulating hormone secretion and the functional activity of the remaining testis in the adult male bonnet monkey (Macaca radiata). Biol Reprod 49(4):743–749
Lunstra DD, Wise TH, Ford JJ (2003) Sertoli cells in the boar testis: changes during development and compensatory hypertrophy after hemicastration at different ages. Biol Reprod 68(1):140–150
Selice R, Ferlin A, Garolla A, Caretta N, Foresta C (2011) Effects of endogenous FSH on normal human spermatogenesis in adults. Int J Androl 34(6 Pt 2):e511-517
Crockford GP, Linger R, Hockley S, Dudakia D, Johnson L, Huddart R, Tucker K, Friedlander M, Phillips KA, Hogg D, Jewett MAS, Lohynska R, Daugaard G, Richard S, Chompret A, Bonaïti-Pellié C, Heidenreich A, Albers P, Olah E, Geczi L, Bodrogi I, Ormiston WJ, Daly PA, Guilford P, Fosså SD, Heimdal K, Tjulandin SA, Liubchenko L, Stoll H, Weber W, Forman D, Oliver T, Einhorn L, McMaster M, Kramer J, Greene MH, Weber BL, Nathanson KL, Cortessis V, Easton DF, Bishop DT, Stratton MR, Rapley EA (2006) Genome-wide linkage screen for testicular germ cell tumour susceptibility loci. Hum Mol Genet 15(3):443–451
Kratz CP, Mai PL, Greene MH (2010) Familial testicular germ cell tumours. Best Pract Res Clin Endocrinol Metab 24(3):503–513
Mai PL, Friedlander M, Tucker K, Phillips KA, Hogg D, Jewett MAS, Lohynska R, Daugaard G, Richard S, Bonaïti-Pellié C, Heidenreich A, Albers P, Bodrogi I, Geczi L, Olah E, Daly PA, Guilford P, Fosså SD, Heimdal K, Liubchenko L, Tjulandin SA, Stoll H, Weber W, Easton DF, Dudakia D, Huddart R, Stratton MR, Einhorn L, Korde L, Nathanson KL, Bishop DT, Rapley EA, Greene MH (2010) The International Testicular Cancer Linkage Consortium: a clinicopathologic descriptive analysis of 461 familial malignant testicular germ cell tumor kindred. Urol Oncol 28(5):492–499
Kelly MP (2014) Does phosphodiesterase 11A (PDE11A) hold promise as a future therapeutic target? Curr Pharm Des 21(3):389–416
Cabanero M, Laje G, Detera-Wadleigh S, McMahon FJ (2009) Association study of phosphodiesterase genes in the sequenced treatment alternatives to relieve depression sample. Pharmacogenet Genomics 19(3):235–238
Delmas V, Laoide B, Masquilier D, de Groot RP, Foulkes NS, Sassone-Corsi P (1992) Alternative usage of initiation codons in CREM generates regulators with opposite functions. Proc Natl Acad Sci USA 89(10):4226–4230
Foulkes NS, Mellstrőm B, Benusiglio E, Sassone-Corsi P (1992) Developmental switch of CREM function during spermatogenesis: from antagonist to activator. Nature 355(6355):80–84
Funding
Open access funding provided by Università degli Studi di Roma La Sapienza within the CRUI-CARE Agreement.. This work was supported by a grant from the Italian Ministry of Education and Research (MIUR-PRIN 2015- 2015XCR88M_006) and “Sapienza” University of Rome, Faculty of Medicine.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
This work was approved by the Policlinico Umberto I—“Sapienza” University of Rome Ethics Committee.
Conflict of interest
The authors declare that there is no conflict of interest.
Human rights
All procedures in this study were performed in accordance with the ethical standards of the institutional research committee and with the 1964 Declaration of Helsinki and subsequent amendments or comparable ethical standards.
Informed consent
Appropriate informed consent was acquired from all participating patients.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Faja, F., Finocchi, F., Carlini, T. et al. PDE11A gene polymorphism in testicular cancer: sperm parameters and hormonal profile. J Endocrinol Invest 44, 2273–2284 (2021). https://doi.org/10.1007/s40618-021-01534-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40618-021-01534-3