Abstract
Purpose
Diabetes is a major public health challenge with widespread prevalence, often leading to complications such as Diabetic Nephropathy (DN)—a chronic condition that progressively impairs kidney function. In this context, it is important to evaluate if Machine learning models can exploit the inherent temporal factor in clinical data to predict the risk of developing DN faster and more accurately than current clinical models.
Methods
Three different databases were used for this literature review: Scopus, Web of Science, and PubMed. Only articles written in English and published between January 2015 and December 2022 were included.
Results
We included 11 studies, from which we discuss a number of algorithms capable of extracting knowledge from clinical data, incorporating dynamic aspects in patient assessment, and exploring their evolution over time. We also present a comparison of the different approaches, their performance, advantages, disadvantages, interpretation, and the value that the time factor can bring to a more successful prediction of diabetic nephropathy.
Conclusion
Our analysis showed that some studies ignored the temporal factor, while others partially exploited it. Greater use of the temporal aspect inherent in Electronic Health Records (EHR) data, together with the integration of omics data, could lead to the development of more reliable and powerful predictive models.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The widespread prevalence of diabetes is still a major public health challenge, with a significant impact on people's quality of life and an increase in mortality. Between 1980 and 2014, the number of people with diabetes increased almost fourfold, from 108 to 422 million, according to the World Health Organization [1]. In the European scenario, 6.2% of adults had diabetes in 2019. Cyprus, Portugal, and Germany were the countries with the highest levels, around 9% or more [2]. In addition, the metabolic control needed to delay diabetes complications is not achieved by the majority of patients. As a result, diabetes can cause many complications, including eye problems (retinopathy), nerve damage (neuropathy), and kidney problems (nephropathy) [3].
Diabetic Nephropathy (DN) is a chronic disease in which the function of the kidneys deteriorates, reducing their ability to eliminate wastes and toxins from the bloodstream and affecting the water balance in the body. DN is considered a progressive disease that usually gets worse over time until the kidneys can no longer function on their own, which is known as end-stage renal disease (ESRD) [4]. It is a disease that is usually considered irreversible although it has been observed that with long-term normalization of the diabetic environment, the architecture of the kidney can undergo significant remodelling and the lesions associated with diabetic nephropathy can be reversed [4]. In developed countries, half of all ESRD cases are due to DN, and the cost of treating ESRD patients is very high [5].
Digitalization has allowed hospitals to store the complete history of patient appointments in a database, resulting in the availability of EHRs. These data are longitudinal because they are collected over time and include multiple patient records at different points in time. Due to the progressive nature of many diseases, a longitudinal approach is usually required to fully assess their development and impact [6]. Given the chronic and long-term nature of diseases such as DN, it is crucial to consider the temporal dimension of patient data and not overlook its importance [7]. The timely implementation of a DN risk assessment may delay or even prevent its progression, which would certainly reduce the number of people with ESRD [8].
The dream of machines that can one day be self-learning without explicit programming is an old one [9]. Machine learning (ML) has its roots in the Artificial Intelligence (AI) movement of the 1950s, with a strong emphasis on practical goals and applications, focusing on tasks such as prediction and optimization [10]. In very simple terms, ML uses various algorithms to learn the patterns and relationships present in a dataset and ultimately predict an outcome. We are now experiencing a major and rapid transformation, brought about by significant advances in ML, which is exponentially increasing automation in many areas of society [11].
ML applied to medicine has great potential to support diagnosis by using a significant amount of patient data and processing it in a fast and intelligent way, helping physicians to make more informed decisions [12]. In fact, ML algorithms can potentially play a crucial role in a faster and more reliable way to diagnose complications associated with diabetes such as DN [13]. The application of ML techniques to analyze EHR data can provide valuable insights and enable the development of ML models that can predict the risk of developing DN or progressing to higher stages, aiding physicians in the diagnosis and ultimately improving the quality of healthcare [14, 15].
There are many studies done on the use of ML to identify cases of diabetic nephropathy. However, the focus of this research is to identify and study the approaches used on clinical EHR data collected over a period of time and the corresponding risk prediction of developing diabetic nephropathy.
This work aims to answer the following research question:
RQ: What are the most effective machine learning techniques used to construct a model that uses the temporal information in diabetic patients' EHR data to predict the development of DN or progression to higher stages?
This literature review was done in a systematic way to ensure that the results are transparent and reproducible, minimizing the bias that would result from the specific choice of studies (cherry-picking) [16].
The main contributions of this work are the following:
-
We present and compare different temporal approaches used in clinical data to develop a predictive model that can accurately identify the risk of developing DN or progressing to higher stages in the future. By providing a comprehensive overview of these approaches, we aim to encourage the development of effective predictive models that can help physicians improve patient outcomes.
-
We contribute to the understanding of the impact that the temporal factor can have on the prediction of DN by reviewing and comparing static and dynamic approaches.
-
We identify the limitations of static and dynamic approaches and highlight the need for further research to improve the accuracy of risk prediction.
-
We show that it is already possible to see that the integration of omics data can potentially improve the results and increase the credibility of predicting DN risk.
The remainder of this paper is organized as follows. Section II describes the methodology used to select the articles to be reviewed. Section III presents the results obtained. A discussion of the main findings arising from these results is presented in Section IV. Threats to the validity of this literature review are presented in Section V, while possible future research directions are outlined in Section VI. Finally, Section VII presents the main conclusions.
Materials and methods
Three databases were used for this literature review: Scopus, Web of Science, and PubMed. These are three of the most popular and reliable sources of scientific information [17]. Only articles written in English and published between January 2015 and December 2022 were included. The search query used was:
“((diabetes) AND ((machine learning) OR (deep learning)) AND ((time) OR (temporal) OR (time series)) AND (predict) AND ((kidney disease) OR (nephropathy)))”.
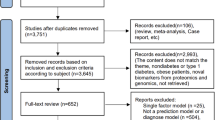
Figure 1 describes the methodology used throughout the process. The first step (Identification) resulted in a total of 164 papers. Based on the references of some of these papers, a further 11 were identified as potentially important, resulting in 175 papers for further analysis. These 11 additional articles were referenced by papers identified in the first stage. During the screening phase, 48 duplicates were removed. In addition, 85 papers were excluded by title and 14 by abstract. These were removed because they did not relate to the intended topic; this phase reduced the original 175 to 28 papers. Of these, only 11 were eligible according to the various criteria defined. Table 1 shows a summary of the excluded articles, the criteria, and a brief explanation of the exclusion criteria.
It should be noted that although the keyword "deep learning" was included in the search query, none of the 11 selected papers used Deep Learning (DL) techniques to solve the problem. With this in mind, we will focus only on approaches that use ML algorithms.
Results
Following the procedure outlined in Fig. 1, 11 articles were included in this review. Artificial intelligence applied to temporal clinical data has the potential to improve the way a diabetic patient is managed according to their risk of developing DN. The different approaches are presented according to different questions: i) which features are most important, ii) what kind of ML models have been created, iii) which ones perform better, and iv) other relevant aspects. The papers selected for this review, together with a summary of their main aspects, are listed in Table 2. Looking at Table 2, we can see that most of the articles were published in the last 2–3 years, which shows a rapid growth in the application of ML to the management of diabetes-related conditions, taking advantage of the large amount of clinical data available.
Data sources
With the emergence and growth of available data, ML models have increased the predictive potential in a wide range of tasks in several application areas. With digitalization, all patient’s data is stored in computer databases. In fact, Electronic Health Records (EHRs) contain vital information about the patient, such as their medical history, illnesses, medications, treatment plans, allergies, and other highly relevant information. This type of data helps clinical research enormously by making it easier to access and track patient data [46]. It also allows for temporal and longitudinal analysis of the data, leading to different approaches and more accurate and correct predictive capabilities [47].
In addition to clinical variables, Omics-based biomarkers are often used. These can be defined as a molecular signature that is identified using omics data and used to predict the presence or risk of a particular disease or condition, or to monitor the response to a particular treatment. Omics can be divided into different research areas such as proteomics (proteins), transcriptomics (RNA), genomics (genes), metabolomics (metabolites), lipidomics (lipids) and epigenomics (methylated DNA) [48].
The integration of omics data with clinical data can significantly improve the ability to analyze and predict complex diseases using ML [49]. The work of Al-Sari et al. [45] is a very good example of the benefits of combining Omics data with clinical data. The performance of some of the models, which had previously been built using only clinical data, increased significantly when Omics data were included. In this case, metabolites, ketones, and sugar derivatives were used. In general, the integration of molecular data will lead to better prognostic models, as demonstrated in several works [50,51,52,53]. Despite the many benefits of integrating this type of data, there are some challenges. Sometimes, even when these data are available, they are very difficult to handle, process, analyze, and finally integrate. This requires specialized knowledge in the branches of mathematics, statistics, biology, and computer science [54].
Feature importance
There are several factors that can lead to the onset or development of DN, such as demographic and genetic factors, clinical measurements, laboratory tests, and medical history. Most of the selected studies used different methods to understand which variables had the greatest influence on the final outcome when predicting risk. Some of these techniques were used to perform feature selection, which can potentially lead to better performance [55].
The work of Chan et al. [42] and Al-Sari et al. [45] used SHapley Additive exPlanations (SHAP) to understand how each feature contributes to the model's predictions, by estimating the amount that each variable contributes to the predicted value of an output. This allows them to ensure that they are selecting the most optimal set of variables for the task.
Recursive Feature Elimination (RFE) is an iterative method that can recursively remove the least important features from a dataset and build a model on the remaining attributes. As presented in Sarkosh et al. [39] and Dong et al. [44], this technique is very useful for selecting a subset of features that aggregates the most important features from a larger dimensional space. In both cases, a variant of this method, Recursive Feature Elimination with Cross-Validation (RFECV), is applied. A very similar approach was adopted by Makino et al. [37] and Dagliatti et al. [36] with their logistic regression (LR) stepwise feature selection method based on the Akaike information criterion (AIC). Stepwise feature selection is a method of selecting a subset of features by iteratively adding or removing variables. The AIC is a trade-off between model goodness and complexity, and measures the relative quality of a statistical model [56]. It can be used in stepwise feature selection to evaluate the performance of the model at each step and decide which feature to add or to remove. Although it appears similar to the RFE method, this technique trains on the selected subset of features at each step and can use either forward selection or backward elimination, whereas RFE trains on all features and removes the least important feature at each step.
Aminian et al. [40] computed the relative importance of each feature in the final model using AIC for the regression models and the Concordance index (C-Index) for the RF models. The C-Index is a metric that considers the temporal dependence associated with the model result and can be used to rank features by importance or even to analyze the global performance of the model.
Singh et al. [35] use a simpler and faster approach, Univariate feature selection, to identify the most relevant variables. These features were chosen through individual statistical tests with the target variable, without considering inter-feature dependencies or relationships.
Song et al. [41] adopted a slightly different approach, using the GBM classifier because it uses an embedded method of feature selection during model training.
Table shows the clinical variables that were mentioned in more than three papers as one of the most prominent variables able to give high predictive power to the model for analyzing the emergence or development of DN, and their respective meaning. Two of the reviewed articles indicated molecular data as being of high importance for the predictive model (Table 3).
Table 4 details the three plasma biomarkers selected by Chan et al., while Table 5 shows the five molecular variables selected by Al-Sari, (2 ketones and 3 sugar derivatives).
Risk models
This section systematizes several approaches to building a model that can predict the risk of developing diabetic nephropathy. Some approaches do not fully exploit the time factor inherent in the data (static approaches), while others manage to make better use of this factor (dynamic/temporal approaches).
Static approaches
Dong et al. [44] used data from non-DN patients at baseline who were followed for three years. The authors then used 408 patients who remained without DN and 408 patients who developed DN after the follow-up period. This data was used to build the model, it contains all the characteristics that the patient presented at baseline and the variable to predict is whether they developed the disease after the three years of follow-up. Binary classification was performed using seven different ML classifiers: Light gradient boosting machine (LightGBM), eXtreme gradient boosting (XGBoost), Adaptive boosting (AdaBoost), Artificial Neural Networks (ANNs), Decision Tree (DT), Support Vector Machine (SVM), and Logistic Regression (LR). This binary classification predicts the presence or absence of DN within 3 years.
There are several other papers that have taken a similar approach and transformed the problem into a binary classification. Romero et al. [38] followed a similar strategy, but defined eight different time windows for all the 7 years of patient follow-up data. Each window corresponds to one year of data, except for the first two windows, which correspond to only 6 months each. The tree-based classifiers OneRule, J48, and RF were chosen for their simplicity, speed of classification, and user-friendly graphical presentation.
Dagliatti et al. [36] used a binary outcome variable but for three different time thresholds of 3, 5, and 7 years to predict the risk of DN. LR, Naïve Bayes (NB), SVM, and RF were tested.
Aminian et al. [40] used data from both surgical and non-surgical patients with T2DM. Multivariate time-to-event regression and RF models were created to predict the 10-year risk of developing DN for both patients with and without metabolic surgery.
Sarkosh et al. [39] trained an LR-based risk score in 1907 diabetic patients, of whom 763 developed DN within five years. In a binary outcome problem, the authors used multivariate LR analysis to generate risk scores and divided patients into four different groups based on their respective risk of DN: low, moderate, high, and very high.
Chan et al. [42] used the same binary outcome in a train/test set of 686 patients and a validation test of 460 patients. Using clinical data and biomarkers, the authors generated risk probabilities using the final RF model and scaled the results to a continuous score between 5 and 100. The authors named the whole system IntelKidneyX. It stratified patients as follows: low risk (46%), intermediate risk (37%) and high risk (17%) of developing DN within 5 years.
Al-Sari et al. [45] and Makino et al. [37] did almost the same as the previously cited papers, but instead of defining outcome as absence or presence, it was defined as progressor or non-progressor in the Al-Sari paper and as worsening or stable in the Makino et al. paper. Al-Sari et al. used data from 190 patients who had no progression of DN and 190 patients who had progression of DN during a mean follow-up of 5.4 years. He used the RF classifier to predict whether the patient would progress to DN during the follow-up period. On the other hand, Makino et al. extracted clinical features from longitudinal, textual, and structural data. LR models were trained using data from 15,422 stable patients (remaining DN stage 1) and 15,388 patients who experienced disease progression at some point (from DN stage 1 to DN stage 2–5).
Unlike the works presented above, Allen et al. [43] are able to predict 3 different outcomes, DN progression to any stage, DN progression to stages 3–5, and DN progression to stages 4–5. Three different models were created for each possible outcome, each predicting the risk of progression to DN over the next 5 years. RF and XGBoost were used as classifiers with a training and test set of 62,994 and 7,656, respectively.
Figure 2 provides a general overview of the different approaches described above.
Dynamic approaches
Different temporal approaches have been proposed to deal with EHR and provide risk prediction for DN. Within the remaining selected articles, the following approaches were used: stacked temporal, multitask temporal, discrete survival, and landmark boosting.
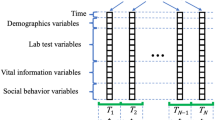
The stacked temporal technique was used in both Singh et al. [35] and Song et al. [41] work. It aggregates data from each time window to create a single prediction. T time windows, with F features in each, result in only one time window with T multiplied by F features. One of the disadvantages of this technique is that the larger the temporal space considered, the higher the dimensionality of the data, which can lead to a large overfitting. In Fig. 3, the physician appointments within each time window are aggregated to form a one-dimensional space, which is then fed into the model and a prediction is obtained.
The multitask temporal method, introduced by Sing et al., involves predicting the outcome separately for each time window, with the requirement that each window must contain at least five physician appointments. When predicting the risk of DN for a new patient, time windows with five or more appointments are used and the final prediction is the average of the different results obtained in each time window. This stratification of the problem is shown in Fig. 4.
Discrete survival and landmark boosting are two techniques mentioned in the paper by Song et al. The first makes an individual prediction in each time window, with no overlap between windows. A disadvantage of this technique is that it assumes that there is no relationship between examples in different time windows, even if they come from the same patient. This can be seen in Fig. 5.
On the other hand, landmark boosting is very similar to discrete survival, but in each time window t, the prediction made in the previous time window t – 1 is also considered. In effect, there is a transfer of knowledge between the time windows, making each prediction more accurate. This can be seen in the representation of the approach shown in Fig. 6, where each model receives not only the features corresponding to a time window, but also the prediction made in the previous time window (Fig. 7).
Used models, interpretation, and performance
This section discusses the type of models most commonly used to predict the onset or development of DN. It also presents the main interpretation techniques used and a comparison of performance.
Considering the selected papers, five different classifiers were proposed: RF, LR, LightGBM, GBM, and Multi-Task Logistic Regression (MTLR). From Fig. 7, we can see that the most selected method was RF, followed by LR, and finally LightGBM, GBM, and MTLR, which were selected only once.
Performance is the most important individual factor that defines the classifier, but it is not the only aspect to consider. RF was the most used classifier because the decision trees that make it up can be interpreted and the final result can be explained [43]. It has a good classification speed and can be represented graphically [38]. However, as a whole, these methods are often difficult to interpret, especially when the number of decision trees is large. It is therefore a classifier with a good balance between speed, complexity, and interpretability. LR has also been proposed several times because it provides a clear interpretation of its coefficients, which are usually represented graphically by nomograms, concepts with which physicians are very familiar [36, 59]. GBM was chosen by Song et al. [41] because of its robustness and effectiveness in predicting DN risk, as demonstrated in previous work. In addition, it incorporates feature selection. MTLR was proposed by Singh et al. [35] because it was appropriate for the type of solution proposed in their multitask temporal methodology. It consists of a multitask learning approach where learning is performed in parallel, and tasks are related to each other [60]. In this case, there is a learning task for each time window, and this approach is used to capture the dependency between tasks.
It is possible to identify three main techniques to interpret the results generated by the predictive model: i) SHapley Additive exPlanations—SHAP values, ii) monograms, iii) decision tree visualization. SHAP values were proposed by Lundberg et al. in 2017 to analyze model predictions [61]. It calculates the importance of each feature for a given prediction, where each feature can have a positive or negative impact on that specific prediction. The contribution of features can be local (each observation) or global (set of observations). In this case, authors used local explanations to show the reasons that lead to a certain result generated by the model for a specific patient. Nomograms are graphical representations of LR models. They work like scoring systems, where each feature is assigned a certain number of points according to its value, and the result varies according to the number of points accumulated in the sum of the different features [62]. Finally, some of the articles used only tree-based models because they can be interpreted directly by visual inspection of the associated decision tree [63].
Some papers predict the onset of DN, some predict the worsening, and some authors predict the worsening for specific stages of the disease. In addition, there are papers where the result corresponds to only one specific time window, while others implement a different prediction for each time window, taking into account a certain number of years. This heterogeneity makes it difficult to compare their performance directly. Table 6 provides detailed information on each of the proposed methods.
Discussion
To the best of our knowledge, this is the first literature review that explores works that make use of EHR data from longitudinally monitored patients to create a predictive DN risk model within a specified time frame. This paper can be used as a basis for further research aimed at in-depth analysis and optimizations on the use of the temporal factor. Such efforts might lead to the development of high-performance predictive models capable of taking advantage of the patient's history to anticipate the onset of diseases such as DN.
There are several approaches in the literature for handling EHR data that are collected over time and then used to build a model to predict the risk of the onset / development of diabetic nephropathy within a given time period. This is a very heterogeneous area of research, where there is no well-defined approach to achieving the previous goal. As Fletcher points out, heterogeneity can be, and usually is, a good thing and can be beneficial [64].
The main findings that have emerged from this work are as follows:
-
There is very little work that takes full advantage of the time factor inherent in EHR data. The works of Sing et al. [35] and Song et al. [41] are an exception. In fact, the landmark boosting method proposed in the Song et al. paper was the approach that took more advantage of the time factor. It not only predicts the risk in each time window, but also takes into account the result produced in the previous time window. Although this approach attempts to exploit the full temporal potential of EHR data, it could still be improved, as it considers all records as independent, which is not the case since each patient has multiple records (appointments).
-
Combining omics data with clinical data can help better predict the risk of DN over time, as confirmed in the work of Al-sari [45]. In the near future, this type of data will be linked to disease risk models because the information they contain is really valuable to increase the predictive power of the different risk models.
-
Another important concern with clinical risk models is interpretability. Almost all of the proposed models were selected not only because of their good performance but also because they allow interpretation of the respective results.
-
The vast majority of the selected articles were published recently (within the last 3 years), demonstrating the importance of studying existing clinical data (EHR) through longitudinal analyses, and the potential that these approaches can have in supporting patient follow-up and medical decision making.
Despite the great capabilities and improvements that these proposed models can potentially bring to medical care, the various papers reviewed have limitations, that are clearly stated by the authors. Some of the most commonly cited limitations are as follows:
-
The patient sample was clinic-based rather than population-based, which means that the model was only tested on a particular dataset, extracted from the population of a particular hospital/clinic. Furthermore, in most studies, there is no external validation dataset, leading to great uncertainty about generalization to a wider population. Cabitza et al. [65] show how external validation is essential for building robust predictive models in medicine.
-
Small data samples, too much missing data and missing important features. Models trained on a small amount of data can result in poor generalizability and lead to incorrect conclusions being drawn. Too much missing data can affect the consistency of the data across different visits by a given patient. This consistency is essential to build a model that can deal with the time factor and make a prediction. In addition, several papers have highlighted various missing demographic, clinical, and laboratory variables that may be essential to improve outcomes.
-
Almost all of the selected papers assume that the examples are independent of each other, which is inaccurate because multiple records belonging to a single patient have been obtained. The ability to account for this inter-record dependency is key to unlocking the potential that may exist in the temporal value of EHR data and can lead to models with greater and better predictive ability. Considering this relationship, Song et al. [41] simulated some inter-record dependency by passing the prediction made in each time window to the prediction of the next time window.
Using the information obtained from the selected articles, we are now able to answer the proposed research question.
RQ: What are the most effective machine learning techniques used to construct a model that uses the temporal information in diabetic patients' EHR data to predict the development of DN or progression to higher stages?
The reviewed literature suggests that despite the potential of using ML techniques to fully exploit the temporal dimension of EHR data to predict the risk of developing or progressing to DN, this has not yet been fully achieved. Many of the techniques used have limited use of the temporal dimension and richness of patient records available in EHR data. Approaches that use only the values available for each patient at baseline or that use statistical operations on the data to combine aggregations of different clinical visits into a single record are valid but completely ignore the temporal potential. There are also some approaches that try to make a longitudinal study of the data, but often in a somewhat incomplete way. For example, the forecasts are separated by time windows (1 year from now, 2 years from now, etc.) and in some cases these forecasts are completely independent of each other. This completely breaks with the value of time and creates a shortcut to a result that is not very different from the first approach. The Landmark Boosting approach proposed by Song et al. was able to stand out because it creates time windows and tries to establish a correlation between these windows by predicting the disease state in the current window based on the state predicted in the previous window.
In summary, all the papers included in this review were generally able to arrive at a workable risk model for the onset or development of DN using a variety of techniques. All of them have attempted, either statically or dynamically, to make partial use of the temporal factor.
Threats to validity
This section discusses all the potential threats to the validation of this work, and the various biases and weaknesses that could in any way jeopardize the results obtained.
This review uses only three different databases, and the search was done with only one query (although it included all relevant keywords). This may introduce a selection bias, meaning that our sample of studies may not be representative of the population studied. If more papers had been included, we would be more likely to have different approaches that could add value to the discussion and possibly change the conclusions drawn.
The heterogeneity of the studies also threatens the validity of this paper. The data differ in quantity, in time of collection, demographic, social and cultural characteristics of the patients, and in some cases even in the meaning of the dependent variable (outcome). Some of them had multiple disease outputs and were not specifically designed to predict the risk of DN. This results in different training and validation data between the different articles selected. They also do not have a standardized way of presenting the results. In addition, some papers omit important information, which can lead to inaccurate or inconsistent results and conclusions. This is commonly referred to as measurement bias.
The study provides a broad and consistent approach to models capable of creating a predictive model of DN using EHR data and their respective time factors. However, it is important to consider that these errors and biases may have altered or influenced the results obtained and the conclusions drawn from them.
Future research directions
Given the small number of works that have been done in this area, there is a great need for future research to have a clearer perspective on the impact that temporal data analysis could have on medical support systems [66]. In the coming years, it is expected that there will be a huge growth in this type of work, as shown by the trends in the studies selected for this review. Therefore, the following future research directions can be outlined:
-
Fully exploit the time factor: Developing strategies that take advantage of the time factor and the dependency between different visits for the same patient, not only to obtain more data, but also to allow the algorithm to access and consider the data as a healthcare team would normally do.
-
ML with omics data: Further and better research into the impact that omics data can have on DN prediction by ML models should be explored so that it is possible to measure the impact of the respective integration. With the advent of modern biotechnologies and the great potential of ML, there is a great opportunity to bring together ML and omics data to significantly improve current systems [67].
-
Apply Deep Learning (DL) techniques: Future research should focus on addressing the temporal nature of EHR data, as most traditional machine learning models are limited in their ability to handle this factor. One promising approach is the use of DL algorithms, which are well suited for detecting hidden patterns in large volumes of data and have greater flexibility and generalizability [68]. Therefore, the application of state-of-the-art DL techniques in future studies could potentially unlock the full temporal potential of EHR data and significantly improve predictive ability.
Conclusion
This review focused on approaches that can use longitudinal data (EHR) to create ML models capable of predicting the risk of onset or development of DN. The findings suggest that the time factor inherent in the data has a clear potential to create a better predictor of DN risk. In addition, the combination of clinical and omics data can help us to achieve better results with greater credibility and generalizability. Furthermore, it is possible to test the concern of the authors of the different papers to create interpretable models whose results can be easily explained and understood by healthcare professionals.
It is important to emphasize that the studies varied in population, type and amount of data, outcome, and even purpose of the study, which may lead to limitations in the findings of this review. Further research is needed to address these limitations and to monitor how this area of temporal analysis of longitudinal data develops in the coming years.
Currently, there are only a few studies that have partially used the temporal information from EHRs to improve the accuracy of predictive ML models. However, we believe that using these temporal data will have a significant impact, especially in the detection of chronic diseases that take a long time to develop symptoms. Physicians use a patient's medical history to diagnose such diseases, and it is important for ML models to do the same. Therefore, incorporating temporal data from EHRs into ML risk prediction models has the potential to be a valuable support tool in healthcare, particularly in the diagnosis and management of chronic diseases, such as DN.
References
“Diabetes.” Accessed: Oct. 29, 2022. [Online]. Available: https://www.who.int/news-room/fact-sheets/detail/diabetes
OECD, Health at a Glance: Europe 2020: State of Health in the EU Cycle. Paris: Organisation for Economic Co-operation and Development, 2020. Accessed: Oct. 29, 2022. [Online]. Available: https://www.oecd-ilibrary.org/social-issues-migration-health/health-at-a-glance-europe-2020_82129230-en
Bloomgarden ZT. Diabetes complications. Diabetes Care. 2004;27(6):1506–14. https://doi.org/10.2337/diacare.27.6.1506.
Fioretto P, Barzon I, Mauer M. Is diabetic nephropathy reversible? Diabetes Res Clin Pract. 2014;104(3):323–8. https://doi.org/10.1016/j.diabres.2014.01.017.
“Diabetic Kidney Disease: A Report From an ADA Consensus Conference | Diabetes Care | American Diabetes Association.” Accessed: Oct. 29, 2022. [Online]. Available: https://diabetesjournals.org/care/article/37/10/2864/30796/Diabetic-Kidney-Disease-A-Report-From-an-ADA
Hund H, Gerth S, Lossnitzer D, Fegeler C. Longitudinal data driven study design. Stud Health Technol Inform. 2014;205:373–377. https://doi.org/10.3233/978-1-61499-432-9-373.
Ponchiardi C, Mauer M, Najafian B. Temporal profile of diabetic nephropathy pathologic changes. Curr Diab Rep. 2013;13(4):592–9. https://doi.org/10.1007/s11892-013-0395-7.
Thomas MC, et al. Diabetic kidney disease. Nat Rev Dis Primer. 2015;1(1):1. https://doi.org/10.1038/nrdp.2015.18.
Kubat M. An Introduction to Machine Learning. Cham: Springer International Publishing; 2021. https://doi.org/10.1007/978-3-030-81935-4.
Bi Q, Goodman KE, Kaminsky J, Lessler J. What is machine learning? a primer for the epidemiologist. Am J Epidemiol. 2019;188(12):2222–39. https://doi.org/10.1093/aje/kwz189.
Brynjolfsson E, Mitchell T. What can machine learning do? workforce implications. Science. 2017;358(6370):1530–4. https://doi.org/10.1126/science.aap8062.
Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019;380(14):1347–58. https://doi.org/10.1056/NEJMra1814259.
Sambyal N, Saini P, Syal R. A review of statistical and machine learning techniques for microvascular complications in type 2 diabetes. Curr Diabetes Rev. 2021;17(2):143–55.
Wong J, Murray Horwitz M, Zhou L, Toh S. Using machine learning to identify health outcomes from electronic health record data. Curr Epidemiol Rep. 2018;5(4):331–42. https://doi.org/10.1007/s40471-018-0165-9.
Kavakiotis I, Tsave O, Salifoglou A, Maglaveras N, Vlahavas I, Chouvarda I. Machine learning and data mining methods in diabetes research. Comput Struct Biotechnol J. 2017;15:104–16. https://doi.org/10.1016/j.csbj.2016.12.005.
Murphy KR, Aguinis H. HARKing: how badly can cherry-picking and question trolling produce bias in published results? J Bus Psychol. 2019;34(1):1–17. https://doi.org/10.1007/s10869-017-9524-7.
Falagas ME, Pitsouni EI, Malietzis GA, Pappas G. Comparison of PubMed, scopus, web of science, and Google scholar: strengths and weaknesses. FASEB J. 2008;22(2):338–42. https://doi.org/10.1096/fj.07-9492LSF.
Song X, Waitman LR, Hu Y, Yu ASL, Robins D, Liu M. Robust clinical marker identification for diabetic kidney disease with ensemble feature selection. J Am Med Inform Assoc. 2019;26(3):242–53. https://doi.org/10.1093/jamia/ocy165.
Connolly P, et al. Analytical validation of a multi-biomarker algorithmic test for prediction of progressive kidney function decline in patients with early-stage kidney disease. Clin Proteomics. 2021;18(1):26. https://doi.org/10.1186/s12014-021-09332-y.
Singh V, Asari VK, Rajasekaran R. A deep neural network for early detection and prediction of chronic kidney disease. Diagnostics. 2022;12(1):116. https://doi.org/10.3390/diagnostics12010116.
Using Machine Learning to Predict Diabetes Complications | IEEE Conference Publication | IEEE Xplore.” Accessed: Dec. 04, 2022. [Online]. Available: https://ieeexplore.ieee.org/document/9677649
Jian Y, Pasquier M, Sagahyroon A, Aloul F. A machine learning approach to predicting diabetes complications. Healthcare. 2021;9(12):1712. https://doi.org/10.3390/healthcare9121712.
David SK, Rafiullah M, Siddiqui K. Comparison of different machine learning techniques to predict diabetic kidney disease. J Healthc Eng. 2022;2022:e7378307. https://doi.org/10.1155/2022/7378307.
Zuo M, Zhang W, Xu Q, Chen D. Deep personal multitask prediction of diabetes complication with attentive interactions predicting diabetes complications by multitask-learning. J Healthc Eng. 2022;2022:5129125. https://doi.org/10.1155/2022/5129125.
Fan Y, Long E, Cai L, Cao Q, Wu X, Tong R. Machine learning approaches to predict risks of diabetic complications and poor glycemic control in nonadherent type 2 diabetes. Front Pharmacol. 2021;12:665951. https://doi.org/10.3389/fphar.2021.665951.
Ravizza S, et al. Predicting the early risk of chronic kidney disease in patients with diabetes using real-world data. Nat Med. 2019;25(1):57–9. https://doi.org/10.1038/s41591-018-0239-8.
Chowdhury NH, et al. Performance analysis of conventional machine learning algorithms for identification of chronic kidney disease in type 1 diabetes mellitus patients. Diagn Basel Switz. 2021;11(12):2267. https://doi.org/10.3390/diagnostics11122267.
Neves AL, et al. Using electronic health records to develop and validate a machine-learning tool to predict type 2 diabetes outcomes: a study protocol. BMJ Open. 2021;11(7):e046716. https://doi.org/10.1136/bmjopen-2020-046716.
Miotto R, Li L, Kidd BA, Dudley JT. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep. 2016;6:26094. https://doi.org/10.1038/srep26094.
Swan BP, Mayorga ME, Ivy JS. The SMART framework: selection of machine learning algorithms with ReplicaTions—A case study on the microvascular complications of diabetes. IEEE J Biomed Health Inform. 2022;26(2):809–17. https://doi.org/10.1109/JBHI.2021.3094777.
Novitski P, Cohen CM, Karasik A, Hodik G, Moskovitch R. Temporal patterns selection for all-cause mortality prediction in T2D with ANNs. J Biomed Inform. 2022;134:104198. https://doi.org/10.1016/j.jbi.2022.104198.
Xu Q, Wang L, Sansgiry SS. A systematic literature review of predicting diabetic retinopathy nephropathy and neuropathy in patients with type 1 diabetes using machine learning. J Med Artif Intell. 2020;3:6. https://doi.org/10.21037/jmai.2019.10.04.
Rahman T, Farzana SM, Khanom AZ. Prediction of diabetes induced complications using different machine learning algorithms. Thesis, BRAC University (2018). Accessed: Dec. 04, 2022. [Online]. Available: http://dspace.bracu.ac.bd/xmlui/handle/10361/10945
Jiang W, et al. Establishment and validation of a risk prediction model for early diabetic kidney disease based on a systematic review and meta-analysis of 20 cohorts. Diabetes Care. 2020;43(4):925–33. https://doi.org/10.2337/dc19-1897.
Singh A, Nadkarni G, Gottesman O, Ellis SB, Bottinger EP, Guttag JV. Incorporating temporal EHR data in predictive models for risk stratification of renal function deterioration. J Biomed Inform. 2015;53:220–8. https://doi.org/10.1016/j.jbi.2014.11.005.
Dagliati A, et al. Machine learning methods to predict diabetes complications. J Diabetes Sci Technol. 2018;12(2):295–302. https://doi.org/10.1177/1932296817706375.
Makino M, et al. Artificial intelligence predicts the progression of diabetic kidney disease using big data machine learning. Sci Rep. 2019;9:11862. https://doi.org/10.1038/s41598-019-48263-5.
Rodriguez-Romero V, Bergstrom RF, Decker BS, Lahu G, Vakilynejad M, Bies RR. Prediction of nephropathy in type 2 diabetes: an analysis of the ACCORD trial applying machine learning techniques. Clin Transl Sci. 2019;12(5):519–28. https://doi.org/10.1111/cts.12647.
Hosseini Sarkhosh SM, Hemmatabadi M, Esteghamati A. Development and validation of a risk score for diabetic kidney disease prediction in type 2 diabetes patients: a machine learning approach. J Endocrinol Invest. 2022;46:415. https://doi.org/10.1007/s40618-022-01919-y.
Aminian A, et al. Predicting 10-year risk of end-organ complications of type 2 diabetes with and without metabolic surgery: a machine learning approach. Diabetes Care. 2020;43(4):852–9. https://doi.org/10.2337/dc19-2057.
Song X, Waitman LR, Yu AS, Robbins DC, Hu Y, Liu M. Longitudinal risk prediction of chronic kidney disease in diabetic patients using a temporal-enhanced gradient boosting machine: retrospective cohort study. JMIR Med Inform. 2020;8(1):e15510. https://doi.org/10.2196/15510.
Chan L, et al. Derivation and validation of a machine learning risk score using biomarker and electronic patient data to predict progression of diabetic kidney disease. Diabetologia. 2021;64(7):1504–15. https://doi.org/10.1007/s00125-021-05444-0.
Allen A, et al. Prediction of diabetic kidney disease with machine learning algorithms, upon the initial diagnosis of type 2 diabetes mellitus. BMJ Open Diabetes Res Care. 2022;10(1):e002560. https://doi.org/10.1136/bmjdrc-2021-002560.
Dong Z, et al. Prediction of 3-year risk of diabetic kidney disease using machine learning based on electronic medical records. J Transl Med. 2022;20(1):143. https://doi.org/10.1186/s12967-022-03339-1.
Al-Sari N, et al. Precision diagnostic approach to predict 5-year risk for microvascular complications in type 1 diabetes. eBioMedicine. 2022;80:104032. https://doi.org/10.1016/j.ebiom.2022.104032.
Cowie MR, et al. Electronic health records to facilitate clinical research. Clin Res Cardiol. 2017;106(1):1–9. https://doi.org/10.1007/s00392-016-1025-6.
Sanchez P, Voisey JP, Xia T, Watson HI, O’Neil AQ, Tsaftaris SA. Causal machine learning for healthcare and precision medicine. R Soc Open Sci. 2022;9(8):220638. https://doi.org/10.1098/rsos.220638.
Micheel CM et al. Omics-Based Clinical Discovery: Science, Technology, and Applications. National Academies Press (US), (2012) Accessed: Jan. 07, 2023. [Online]. Available: https://www.ncbi.nlm.nih.gov/books/NBK202165/
Holzinger A, Haibe-Kains B, Jurisica I. Why imaging data alone is not enough: AI-based integration of imaging, omics, and clinical data. Eur J Nucl Med Mol Imaging. 2019;46(13):2722–30. https://doi.org/10.1007/s00259-019-04382-9.
Jayawardana K, et al. Determination of prognosis in metastatic melanoma through integration of clinico-pathologic, mutation, mRNA, microRNA, and protein information. Int J Cancer. 2015;136(4):863–74. https://doi.org/10.1002/ijc.29047.
Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res. 2018;24(6):1248–59. https://doi.org/10.1158/1078-0432.CCR-17-0853.
Tong D, et al. Improving prediction performance of colon cancer prognosis based on the integration of clinical and multi-omics data. BMC Med Inform Decis Mak. 2020;20(1):22. https://doi.org/10.1186/s12911-020-1043-1.
De Bin R, Sauerbrei W, Boulesteix A-L. Investigating the prediction ability of survival models based on both clinical and omics data: two case studies. Stat Med. 2014;33(30):5310–29. https://doi.org/10.1002/sim.6246.
Cambiaghi A, Ferrario M, Masseroli M. Analysis of metabolomic data: tools, current strategies and future challenges for omics data integration. Brief Bioinform. 2017;18(3):498–510. https://doi.org/10.1093/bib/bbw031.
Cai J, Luo J, Wang S, Yang S. Feature selection in machine learning: a new perspective. Neurocomputing. 2018;300:70–9. https://doi.org/10.1016/j.neucom.2017.11.077.
Cavanaugh JE, Neath AA. The akaike information criterion: background, derivation, properties, application, interpretation, and refinements. WIREs Comput Stat. 2019;11(3):e1460. https://doi.org/10.1002/wics.1460.
Wajant H, Siegmund D. TNFR1 and TNFR2 in the control of the life and death balance of macrophages. Front Cell Dev Biol. 2019;7:91. https://doi.org/10.3389/fcell.2019.00091.
Tanase DM, et al. The predictive role of the biomarker kidney Molecule-1 (KIM-1) in acute kidney injury (AKI) cisplatin-induced nephrotoxicity. Int J Mol Sci. 2019;20(20):5238. https://doi.org/10.3390/ijms20205238.
Jiang S, et al. Prognostic nomogram and score to predict renal survival of patients with biopsy-proven diabetic nephropathy. Diabetes Res Clin Pract. 2019;155:107809. https://doi.org/10.1016/j.diabres.2019.107809.
Thung K-H, Wee C-Y. A brief review on multi-task learning. Multimed Tools Appl. 2018;77(22):29705–25. https://doi.org/10.1007/s11042-018-6463-x.
Lundberg SM, Lee S-I. A Unified Approach to Interpreting Model Predictions, in Advances in Neural Information Processing Systems. Curran Associates, Inc (2017);. Accessed: Jun. 13, 2022. [Online]. Available: https://proceedings.neurips.cc/paper/2017/hash/8a20a8621978632d76c43dfd28b67767-Abstract.html
Salkind N. Encyclopedia of Research Design. SAGE Publications, Inc. 2010. https://doi.org/10.4135/9781412961288
Zhao X, Wu Y, Lee DL, Cui W. iForest: interpreting random forests via visual analytics. IEEE Trans Vis Comput Graph. 2019;25(1):407–16. https://doi.org/10.1109/TVCG.2018.2864475.
Fletcher J. What is heterogeneity and is it important? BMJ. 2007;334(7584):94–6. https://doi.org/10.1136/bmj.39057.406644.68.
Cabitza F, et al. The importance of being external. methodological insights for the external validation of machine learning models in medicine. Comput Methods Programs Biomed. 2021;208:106288. https://doi.org/10.1016/j.cmpb.2021.106288.
Bica I, Alaa AM, Lambert C, van der Schaar M. From real-world patient data to individualized treatment effects using machine learning: current and future methods to address underlying challenges. Clin Pharmacol Ther. 2021;109(1):87–100. https://doi.org/10.1002/cpt.1907.
Li R, Li L, Xu Y, Yang J. Machine learning meets omics: applications and perspectives. Brief Bioinform. 2022;23(1):1–22. https://doi.org/10.1093/bib/bbab460.
Xie F, et al. Deep learning for temporal data representation in electronic health records: a systematic review of challenges and methodologies. J Biomed Inform. 2022;126:103980. https://doi.org/10.1016/j.jbi.2021.103980.
Funding
Open access funding provided by FCT|FCCN (b-on).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mesquita, F., Bernardino, J., Henriques, J. et al. Machine learning techniques to predict the risk of developing diabetic nephropathy: a literature review. J Diabetes Metab Disord (2023). https://doi.org/10.1007/s40200-023-01357-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40200-023-01357-4