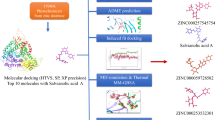
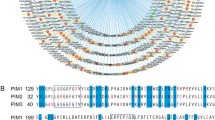
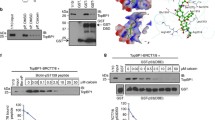
Abstract
Cancer is a serious health concern growing at a rapid speed where normal cells take neoplastic transformation. Different pathway is tightly regulated with each other to maintain the harmony and sudden changes in single protein leads to aberrant changes in the whole system. Development of drugs to target these proteins aimed to block the signaling route that leads to cell death. Here, in this study, we performed in silico expression analysis of these potential proteins using system biological approach by mimicking the cell and understanding the behavior of different proteins in drugging condition. We performed in silico biomolecular interaction analysis for exploring the potential plant-derived compounds that can be served as an anticancerous drug with least toxicity by comparing with reference drug approved by FDA. Our results suggest that PI3K, p53-Mdm2 proteins are ideal proteins for targeting cancer cells, while overexpression of mTOR protein was observed when drug targeted this receptor. We state that PI3K family protein plays important role in drug discovery, and compounds obtained from in silico analysis can be served as a potential anticancerous drug for treating different cancer types.









Similar content being viewed by others
References
Anderson AR, Quaranta V (2008) Integrative mathematical oncology. Nat Rev Cancer 8(3):227–234
Arora R, Gupta D, Chawla R, Sagar R, Sharma A, Kumar R, Prasad J, Singh S, Samanta N, Sharma RK (2005) Radioprotection by plant products: present status and future prospects. Phytother Res 19(1):1–22
Arora D, Singh V, Singh A (2016) Modeling and simulation analysis of Salmonella typhimurium inside human epithelial cells: host-pathogen relationship analysis by system biology. In: Bioinformatics and systems biology (BSB), IEEE, pp 1–4
Arora D, Chaudhary R, Singh A (2017a) System biology approach to identify potential receptor for targeting cancer and biomolecular interaction studies of indole [2, 1-a] isoquinoline derivative as anticancerous drug candidate against it. Interdiscip Sci Sciences 26:1
Arora D, Jyoti K, Singh A (2017b) Understanding the role of Salmonella pathogenic island 1 (SPI-I) and host-pathogen interaction for typhoid using system biology approach. Int J Bioinform Res Appl 13(3):187–199
Bader AG, Kang S, Vogt PK (2006) Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci 103(5):1475–1479
Beauchamp EM, Platanias LC (2013) The evolution of the TOR pathway and its role in cancer. Oncogene 32(34):3923–3932
Boyd MR, Paull KD (1995) Some practical considerations and applications of the National Cancer Institute in vitro anticancer drug discovery screen. Drug Dev Res 34(2):91–109
Brent R (2000) Genomic biology. Cell 100(1):169–183
Chène P (2003) Inhibiting the p53–MDM2 interaction: an important target for cancer therapy. Nat Rev Cancer 3(2):102–109
Chico LK, Van Eldik LJ, Watterson DM (2009) Targeting protein kinases in central nervous system disorders. Nat Rev Drug Discov 8(11):892–909
Clarke PA, te Poele R, Wooster R, Workman P (2001) Gene expression microarray analysis in cancer biology, pharmacology, and drug development: progress and potential. Biochem Pharmacol 62(10):1311–1336
Cohen P (2002) Protein kinases—the major drug targets of the twenty-first century? Nat Rev Drug Discov 1(4):309–315
Cohen P, Alessi DR (2012) Kinase drug discovery–what’s next in the field? ACS Chem Biol 8(1):96–104
Csizmadia P (1999) September. MarvinSketch and MarvinView: molecule applets for the World Wide Web. In: Proceedings of ECSOC-3, the third international electronic conference on synthetic organic chemistry, September 1ą30, pp 367–369
Dräger A, Hassis N, Supper J, Schröder A, Zell A (2008) SBMLsqueezer: a cell designer plug-into generate kinetic rate equations for biochemical networks. BMC Syst Biol 2(1):39
Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ (2016) Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc 11(5):905–919
Fridman JS, Lowe SW (2003) Control of apoptosis by p53. Oncogene 22(56):9030–9040
Funahashi A, Morohashi M, Kitano H, Tanimura N (2003) Cell Designer: a process diagram editor for gene-regulatory and biochemical networks. Biosilico 1(5):159–162
Funahashi A, Morohashi M, Matsuoka Y, Jouraku A, Kitano H (2007) cell designer: a graphical biological network editor and workbench interfacing simulator. In: Choi S (ed) Introduction to systems biology. Humana Press, New York, pp 422–434
Funahashi A, Matsuoka Y, Jouraku A, Morohashi M, Kikuchi N, Kitano H (2008) Cell Designer 3.5: a versatile modeling tool for biochemical networks. Proc IEEE 96(8):1254–1265
Gressner AM, Weiskirchen R (2006) Modern pathogenetic concepts of liver fibrosis suggest stellate cells and TGF-β as major players and therapeutic targets. J Cell Mol Med 10(1):76–99
Hainaut P, Hollstein M (1999) p53 and human cancer: the first ten thousand mutations. Adv Cancer Res 77:81–137
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674
Hartman JL, Garvik B, Hartwell L (2001) Principles for the buffering of genetic variation. Science 291(5506):1001–1004
Hendriks BS, Hua F, Chabot JR (2008) Analysis of mechanistic pathway models in drug discovery: p38 pathway. Biotechnol Prog 24(1):96–109
Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U (2006) COPASI—a complex pathway simulator. Bioinformatics 22(24):3067–3074
Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D (2003) Cornish-Bowden A, Cuellar AA. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19(4):524–531
Hutchinson TE, Zhang J, Xia SL, Kuchibhotla S, Block ER, Patel JM (2012) Enhanced phosphorylation of caveolar PKC-α limits peptide internalization in lung endothelial cells. Mol Cell Biochem 360(1–2):309–320
Insall RH, Weiner OD (2001) PIP3, PIP2, and cell movement—similar messages, different meanings? Dev Cell 1(6):743–747
Jain RK, Duda DG, Clark JW, Loeffler JS (2006) Lessons from phase III clinical trials on anti-VEGF therapy for cancer. Nat Clin Pract Oncol 3(1):24–40
Jänne PA, Gray N, Settleman J (2009) Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nat Rev Drug Discov 8(9):709–723
Jayaraj P, Sen S, Sharma A, Chosdol K, Kashyap S, Rai A, Pushker N, Bajaj M (2015) Eyelid sebaceous carcinoma: a novel mutation in lymphoid enhancer-binding factor-1. Br J Dermatol 173(3):811–814
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30
Koutsogiannouli E, Papavassiliou AG, Papanikolaou NA (2013) Complexity in cancer biology: is systems biology the answer? Cancer Med 2(2):164–177
Kreeger PK, Lauffenburger DA (2010) Cancer systems biology: a network modeling perspective. Carcinogenesis 31(1):2–8
Lakin ND, Jackson SP (1999) Regulation of p53 in response to DNA damage. Oncogene 18(53):7644–7655
Laplante M, Sabatini DM (2009) mTOR signaling at a glance. J Cell Sci 122(20):3589–3594
Lewis NE, Abdel-Haleem AM (2013) The evolution of genome-scale models of cancer metabolism. Front Physiol 4:237
Lin MC, Lin GZ, Hwang CI, Jian SY, Lin J, Shen YF, Lin G (2012) Synthesis and evaluation of a new series of tri-, di-, and mono-N-alkylcarbamylphloroglucinols as conformationally constrained inhibitors of cholesterol esterase. Protein Sci 21(9):1344–1357
Melnikova I, Golden J (2004) Targeting protein kinases. Nat Rev Drug Discov 3(12):993–994
Momand J, Wu HH, Dasgupta G (2000) MDM2—master regulator of the p53 tumor suppressor protein. Gene 242(1):15–29
Prives C (1998) Signaling to p53: breaking the MDM2–p53 circuit. Cell 95(1):5–8
Rozengurt E, Soares HP, Sinnet-Smith J (2014) Suppression of feedback loops mediated by PI3K/mTOR induces multiple over activation of compensatory pathways: an unintended consequence leading to drug resistance. Mol Cancer Ther 13(11):2477–2488
Sakaguchi K, Herrera JE, Saito SI, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E (1998) DNA damage activates p53 through a phosphorylation–acetylation cascade. Genes Dev 12(18):2831–2841
Sawyers CL, Abate-Shen C, Anderson KC, Barker A, Baselga J, Berger NA, Foti M, Jemal A, Lawrence TS, Li CI, Mardis ER (2013) AACR cancer progress report 2013. Clin Cancer Res 19(20 Suppl):S1–S98
Shangary S, Wang S (2008) Targeting the MDM2-p53 interaction for cancer therapy. Clin Cancer Res 14(17):5318–5324
Shi D, Gu W (2012) Dual roles of MDM2 in the regulation of p53 ubiquitination dependent and ubiquitination independent mechanisms of MDM2 repression of p53 activity. Genes Cancer 3(3–4):240–248
Singh L, Pushker N, Sen S, Singh MK, Chauhan FA, Kashyap S (2015) Prognostic significance of polo-like kinases in retinoblastoma: correlation with patient outcome, clinical and histopathological parameters. Clin Exp Ophthalmol 43(6):550–557
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461
Turkson J, Jove R (2000) STAT proteins: novel molecular targets for cancer drug discovery. Oncogene 19(56):6613
Vogelstein B, Lane D, Levine AJ (2000a) Surfing the p53 network. Nature 408(6810):307–310
Vogelstein B, Lane D, Levine AJ (2000b) Surfing the p53 network. Nature 408(6810):307–310
von Manstein V, Min Yang C, Richter D, Delis N, Vafaizadeh V, Groner B (2013) Resistance of cancer cells to targeted therapies through the activation of compensating signaling loops. Curr Signal Transduct Ther 8(3):193–202
Vousden KH, Lu X (2002) Live or let die: the cell’s response to p53. Nat Rev Cancer 2(8):594–604
Wade M, Li YC, Wahl GM (2013) MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 13(2):83–96
Yang Y, Li CC, Weissman AM (2004) Regulating the p53 system through ubiquitination. Oncogene 23(11):2096–2106
Yarden Y, Pines G (2012) The ERBB network: at last, cancer therapy meets systems biology. Nat Rev Cancer 12(8):553–563
Yıldırım MA, Goh KI, Cusick ME, Barabási AL, Vidal M (2007) Drug—target network. Nat Biotechnol 25(10):1119–1126
Acknowledgement
This study was conducted in the Department of Biotechnology, G.B. Pant Engineering College (GBPEC), Pauri Garhwal (Uttarakhand). Devender Arora is thankful to TEQIP-II (Technical Education Quality Improvement Program) for financial assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors have no conflict of interest regarding the publication of paper.
Rights and permissions
About this article
Cite this article
Arora, D., Singh, A. Systems biology approach deciphering the biochemical signaling pathway and pharmacokinetic study of PI3K/mTOR/p53-Mdm2 module involved in neoplastic transformation. Netw Model Anal Health Inform Bioinforma 7, 2 (2018). https://doi.org/10.1007/s13721-017-0162-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-017-0162-9