Abstract
The precise quantification of microRNAs (miRNAs) expression level is a critical factor in mastering its functions. We evaluate the suitability of two common genes and ten miRNAs as normalizers for miRNA quantification in the head and ovary at different reproductive status of bumblebees, Bombus lantschouensis by using four different algorithms and one consensus rank approach. For the head and ovary combination, miR-275 was the best candidate. For different tissues, miR-275 was the most stable candidate in the head, while the candidate for the ovary was miR-277. To test the best candidate accuracy, miR-315 was demonstrated to be downregulated based on miR-275 normalization in ovipositor bumblebees. The miR-275 and miR-277 combination is identified to be the most reliable and suitable reference genes for the head and ovary of bumblebees.
Similar content being viewed by others
1 Introduction
Bumblebees are one of the most efficient pollinators for many plants, especially in alpine ecosystem (Williams et al. 2015). In crops, bumblebee pollination can increase fruit quality and yield significantly. Since the 1990s, bumblebees have been introduced to greenhouses for crop pollination and brought a large increase in economic income from the pollination services. Therefore, the commercial production of bumblebee colonies for crop pollination promotes the development of artificial rearing all over the world (Velthuis and van Doorn 2006). The fecundity of the queen is one of the vital factors that primarily affect the large-scale production of bumblebees. The food quality, queen quality, and environmental factors that influence the egg laying of the queens and success of colony foundation were explored (Yeninar et al. 2000). However, there are many questions regarding the genetic basis of fecundity regulation that have not been answered to date.
MicroRNAs (miRNAs) represent a family of small (19–24 nucleotides), single-stranded, non-coding, endogenous RNA molecules (Bartel 2009). The important function of small RNAs is involved in the transcriptional and post-transcriptional regulation of gene expression related to different biological processes (Marco et al. 2013). It has been demonstrated that miRNAs regulate reproductive function of the ovary (Baley and Li 2012). Moreover, it can switch the reproductive mode and development (Quah et al. 2015). Recent research showed that the expression levels of miRNAs were associated with honeybee diverse biological functions, such as ovarian activity, caste differentiation, and foraging behavior (Li et al. 2012; Guo et al. 2016; Macedo et al. 2016). In bumblebees, miRNAs have been described as an important gene in the regulation of bumblebee queen egg-laying (Sun et al. 2015). Therefore, miRNAs are key factors for regulating reproductive status of the queen in year-round rearing of bumblebees.
Quantitative real-time PCR (RT-qPCR) is a gold standard technique for measuring miRNA expression that is widely used because of its sensitivity, large dynamic range, and accurate quantification (Huggett et al. 2005; Kang et al. 2012; Li et al. 2012). To reduce the sample and experimental variation, this type of analysis requires a normalization of raw expression data (Huggett et al. 2005). The adoption of reference genes was the most convenient and popular approach for normalizing raw expression data. However, Tichopad et al. (2004) observed that unknown tissue-specific factors can influence amplification kinetics, but this effect can be ameliorated by appropriate primer selection. Therefore, in using RT-qPCR to correctly evaluate expression levels of miRNAs to explore their function, an ideal reference gene is required. These reference genes should be adequately expressed in the tissue of interest with minimal variability under different conditions and samples (Dheda et al. 2004; Huggett et al. 2005). Therefore, the screening and validation of reference genes is a crucial and necessary step for miRNA expression measurement. How does one select a suitable reference gene? The different algorithms such as delta-Ct (Silver et al. 2006), Normfinder (Andersen et al. 2004), geNorm (Vandesompele et al. 2002), and BestKeeper (Pfaffl et al. 2004) were developed to rank the candidate reference genes according to their stability and optimal combined numbers. Until recently, there are no reports about specific recommendations of a suitable and stable reference gene for miRNA quantification in social insects, such as honeybees and bumblebees, under different reproductive status.
Bombus lantschouensis Vogt was one of the bumblebee species that was selected for artificial rearing and greenhouse crop pollination in China. These bees were widely distributed at medium elevation mountains and plateaus within North China (An et al. 2014). Moreover, a large size of colonies, numbers of virgins, and tame temper make them an excellent species for artificial rearing. Recent evidence indicates that the pollination efficiency of this species is outperforming compared with the honeybee. The interesting fact regarding B. lantschouensis is that it visits flowers with less pollen and nectar than Apis mellifera (Zhou et al. 2015). Considering such pollinating behavior, B. lantschouensis became one of the most selective species for large-scale rearing in China. Other reproductive behaviors such as the queen’s oviposition regulation are a crucial aspect to artificial rearing on an industrial scale. Sun et al. (2015) explored the differential expression of miRNAs between egg-laying and non-egg-laying queen’s head of B. lantschouensis. However, there is no documentation regarding the reference gene selection and validation for miRNA quantification in the head and ovary of the bumblebee queen. Therefore, it is necessary to test their stability in bumblebees in order to obtain the correct miRNA expression level.
The aim of the present study was designed to evaluate the suitability of potential reference genes for normalizing the expression levels of miRNA in the head and ovary of B. lantschouensis under the different reproductive status. Potential reference genes, including two commonly used reference genes and ten novel potential miRNAs, were compared and ranked with four statistical algorithms and one rank aggregation approach. Finally, the stability of the best candidate was validated by measuring the expression patterns of miR-315, which was reported to regulate the oviposition in bumblebee.
2 Material and methods
2.1 Bees
Queens of B. lantschouensis were collected from the bumblebee rearing room at the Institute of Apicultural Research, Chinese Academy of Agricultural Sciences. Queens were reared in small boxes (18 × 18 × 18 cm). Sucrose solution (50% w/vol) and clumps of honeybee pollen pellets were provided ad libitum every 2 days. After 1 week of the queen laying eggs, heads were cut and ovaries were dissected from three egg-laying and three non-egg-laying queens separately. Each head and each pair of ovary were kept in the Eppendorf tubes independently. In total, 12 tubes were collected included six heads (three egg-laying and three non-egg-laying queens’ heads) and six ovaries (three egg-laying and three non-egg-laying queens’ ovaries). Total RNA was extracted separately from every head and every pair of ovaries. The ovaries were checked to ensure the queen-laying status under a microscope. All of the tissues were kept at − 80 °C until use.
2.2 RNA isolation and cDNA synthesis
Total RNA was extracted from heads and ovaries using the miRcute miRNA Isolation kit (Tiangen, China) according to the manufacturer’s protocol. The quality and quantity of total RNA were determined using denaturing gel electrophoresis and a Nanodrop spectrophotometer (ND-1000, USA), respectively. cDNA synthesis was performed by two methods. For protein-coding genes, reverse transcription was done with random primer method using the PrimeScript™ First-Strand cDNA Synthesis Kit (TaKaRa, Japan). For miRNAs, reverse transcription was done using the 3′ end poly (A) tail binding method of miRcute miRNA First-Strand cDNA kit (Tiangen, China) according to the manufacturer’s protocol. Poly (A) tail was added to mature miRNAs 3′ ends, and then Oligo(dT)-universal tag primer was used to generate the first strand.
2.3 Primers of candidate reference genes
Twelve candidate reference genes including two common reference genes (ACTB and GAPDH) (Hornakova et al. 2010) and ten miRNAs were selected as candidate qPCR normalizers (Table I). Primers were designed using Primer 3 software (Rozen and Skaletsky 2000). The miRNA primer design followed the requirements of miRcute miRNA First-Strand cDNA kit. Those miRNAs were reported to have a stable expression in the head of egg-laying and non-egg-laying queens according to NGS small RNA sequencing (Sun et al., 2015). The sequences of mature miRNAs were listed in Table S1. All primers amplified the candidate reference genes successfully.
2.4 Real-time quantitative PCR
Real-time quantitative PCR was performed in a Stratagene MX3000p amplifier using a special miRNA amplification kit, the miRcute miRNA qPCR Detection kit (Tiangen, China). To obtain primer amplification efficiency, standards of 10-fold dilution samples were run and analyzed for each candidate reference gene. The volume of reaction mixture was carried out according to the kit instruction. Briefly, 0.4 μl of 10 μM miRNA specific forward and 0.4 μl of 10 μM reverse primer, 10 μl of 2× miRcute Plus miRNA Premix (with SYBR&ROX) and 1.6 μl of 50× ROX Reference Dye, 2.0 μl of cDNA, and RNAse-free H2O were added to a final volume 20 μl. PCR amplification conditions were as follows: 2 min at 95 °C followed by 40 cycles of 95 °C for 20 s, and 60 °C for 35 s. A thermal denaturing protocol with a gradient (0.5 °C every 30 s) from 65 to 95 °C formed the dissociation curves and that was used to investigate the specificity of the qPCR reaction and the presence of primer dimers. The melting curve was checked to ensure a single amplicon. Each sample was run in triplicate and the mean cycle threshold (Ct)/fluorescence value was taken for further analysis.
2.5 Data analysis
Real-time PCR data of the twelve candidate reference genes were analyzed by four different algorithms: delta-Ct (Silver et al. 2006), Normfinder (Andersen et al. 2004), geNorm (Vandesompele et al. 2002), and BestKeeper (Pfaffl et al. 2004) using the online tool RefFinder (http://fulxie.0fees.us/?i=2, accessed 2017) (Xie et al. 2012). Delta-Ct determines the stable gene using a simple ΔCt method based on comparing pairs of genes. Bestkeeper selects the suitable gene using geometric mean and repeated pairwise correlation analysis. geNorm calculates the gene expression stability value according to the mean pairwise variation between an individual gene and all other tested control genes. NormFinder assesses the gene stability judging by a mathematical model of gene expression. Based on the stability order determined by four algorithms, the R-package RankAggreg (Version 0.5) was used to access the consensus rank of candidate reference genes (Pihur et al., 2009).
2.6 Reference gene validation
To test the best reference gene for the head and ovary miRNA expression normalization, miR-315 was used as a target gene to measure the transcript levels. This miRNA was demonstrated by RNA-seq to be downregulated in the head of bumblebee queens after oviposition (Sun et al. 2015). The relative quantity of miR-315 was calculated by the double delta-Ct method (Livak and Schmittgen 2001). Heads and ovaries were dissected from three egg-laying and three non-egg-laying queens, and then miRNAs were extracted separately from every head and every pair of ovaries to receive six independent biological samples for each tissue. The difference of expression level was analyzed using ANOVA with the basic package of R-project (Version 3.32).
3 Results
3.1 Expression levels of reference genes
All primers used to amplify the candidate reference genes generated single amplicons, as demonstrated by the single-peak melting curves of the PCR products (Fig. S1). The standard curves were generated for each candidate reference gene using serial dilutions of cDNA from the mixture of head and ovary. The efficiency values of all of the candidate reference genes ranged from 92.10 to 109.35% based on the slopes of the standard curves. The linear correlation coefficients (r) were between 0.978 and 1.000 (Table I). In addition, the single peak of the melting curve for each candidate reference gene confirmed that the amplicon was a product of gene-specific amplification.
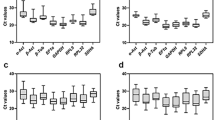
We observed that the Ct values varied in a relatively wide range for each candidate reference gene between head and ovary. The Ct value ranged from 16 to 35 cycles in the head and 15 to 27 cycles in the ovary as represented by box-and-whiskers plots (Figure 1). GAPD gene was the most abundantly expressed gene in the head with the largest variable Ct value. However, the Ct value of miR-14 was the highest expression level and miR-275 was the lowest variability in the ovary. The successful amplification indicates that all the candidate reference genes can be used for the stability analysis.
Expression levels of candidate reference genes in the head and ovary samples of bumblebee queens. a Box-and-whisker plot of the Ct values of ACTB, GAPDH, and ten candidate reference miRNAs (miR-1, miR-2, miR-14, miR-184, miR-275, miR-276, miR-317, miR-9a, miR-7, and miR-277) in head samples. b Box-and-whisker plot of the Ct values of candidate reference genes in ovary samples. Whiskers represent the maximum and minimum values. The median is depicted by the line across the box.
3.2 Most stable expression of candidate reference gene
Four different approaches have been implemented to obtain the expression stability order of candidate reference genes under different reproductive status. Results showed that the stability ranking of candidate reference accessed by four approaches was different in the head or ovary (Table S2). The delta-Ct analysis showed that the standard deviations of Ct values ranged from 1.73 to 6.37 in the group of head and ovary, where miR-184 and miR-317 had the lowest stability value of genes. In the head, the standard deviations of Ct values varied from 0.50 to 1.01. In the ovary, the standard deviations of Ct values increased from 0.72 to 2.11. Therefore, based on the standard deviations of Ct values, the most stable genes were miR-7 and miR-277 in the head and in the ovary, respectively. BestKeeper analysis revealed that ACTB had the lowest standard deviation across the head and ovary under different status. In the head, miR-275 had the most stable expression level. However, miR-7 was the most stable gene expressed in the ovary. Gene stability calculated by GeNorm showed that miR-275 and miR-317 had the most stable expression through the head and ovary (M = 0.397). The most stable genes expressed in the head were miR-14 and miR-276. In the ovary, miR-317 and miR-277 were selected as the most stable candidate reference genes. Normfinder analysis found miR-275 to be the gene with the most stable expression with a stability value of 0.199 across the head and ovary. The miR-275 was also selected as the gene with the most stable expression in the head. In the ovary, miR-14 was the most stable gene.
3.3 Ranking and combining stability of candidate reference genes
The different orders of candidate reference genes estimated by the four algorithms were combined by Rankaggreg R-package to obtain the consensus ranking of reference genes. The means of the cross-entropy Monte Carlo algorithm ranking determined that miR-275 showed the most stable gene expression under the different reproductive status across the head and ovary (Fig. S2). In addition, it was found that miR-275 was also the best stability reference gene in the head. However, miR-277 was selected as the most stable gene in the ovary (Table II).
The optimal number of reference genes was determined by pairwise variations value of normalization factors in geNorm. The results showed that all pairwise variation values were < 0.15 across the head and ovary (Figure 2). When the pairwise variation value is < 0.15, it suggested that no other genes are needed to be added to improve the stability of reference genes. Therefore, based on the pairwise variation value, the combination of two reference genes was enough for normalization under different reproductive status across the head and ovary.
Determination of the optimal number of reference genes for miRNA normalization in head and ovary samples of bumblebee queens by using geNorm.
Pairwise variation (Vn/n+1) analysis between the normalization factors NFn and NFn+1 to determine the number of control genes required for accurate normalization.
3.4 Validation of the candidate reference gene by miR-315 expression
To test the validation of the best candidate reference genes across the head and ovary, miR-315 served as the target gene for analyzing expression levels under different reproductive status. Normalizing the expression values of miR-315 by the candidate reference gene miR-275 showed that the expression level of miR-315 decreased not only in the head, but also in the ovary after queen egg-laying (Figure 3). The expression level of miR-315 under different reproductive status reached a significant difference (head: F1, 16 = 28.643; p < 0.05, ovary: F1, 16 = 391.405; p < 0.001). This result demonstrated that the optimal reference gene can be used to normalize the miRNA expression under different reproductive status.
Effect of normalization on miRNA expression in the head (a) and ovary (b) of queens. Reproductive status differences of queens are observed in miR-315 expression when data are normalized using the candidate reference gene miR-275. There are six independent biological samples for each tissue (n = 6), and three biological replicates for each of the two different reproductive status of female bumblebees. Significant differences of miR-315 expression levels between egg-laying and non-egg-laying queens were determined using one-way ANOVA test.
4 Discussion
With the development of next-generation sequencing, increasing numbers of miRNAs were identified and characterized from non-model organisms. It was demonstrated that miRNAs play an important role in regulating the biological and pathological processes by binding to the mRNA sequences of target genes at the stage of transcription and post-transcription (Plasterk 2006; King et al. 2011; Guo et al. 2016; Macedo et al. 2016). As the content of miRNAs is approximately 0.01–0.1% of the total RNA present in the tissue, it is critical for the accurate measurement of those low abundance miRNAs to master its functions. Currently, qPCR is one of the best methods to detect the subtle expression difference of miRNAs. To obtain reliable and accurate qPCR data, a suitable reference gene should be validated to avoid the erroneous qualification. In this study, we determined the most appropriate reference gene among 12 candidates in normalizing the miRNA of the queen’s head and ovary at the different reproductive status of B. lantschouensis. To eliminate the effect of RNA extraction, reverse transcription, and real-time PCR, a commercial kit of miRNA tailing extending poly(A) method and standard real-time qPCR were conducted for all of the samples in our study.
An accurate and optimized qPCR assay should have a linear correlation coefficient (R2) and a high PCR amplification efficiency from 90 to 110% (Serafin et al. 2014). Our primers tested showed that they reached the requirement of linear correlation and PCR amplification efficiency (Table II). Therefore, all of the primers are suitable for the stability validation. The expression level of different candidate reference genes was notably variable (Figure 1). From the Ct value, we observed that the variation is larger in inter-tissues than in intra-tissues for most of candidate reference genes under the different reproductive status. Moreover, the change in Ct value is higher among tissues than among reproductive status. Therefore, we suggested that we should test the stability of reference genes in the same tissue under different reproductive status and use the most stable candidate reference gene independently.
To obtain the suitable reference gene, multiple algorithms were developed to analyze the qPCR data and make the conclusion more reliable (Vandesompele et al. 2002; Andersen et al. 2004; Pfaffl et al. 2004; Silver et al. 2006). Four approaches (geNorm, NormFinder, BestKeeper, and delta-Ct) were used to evaluate our qPCR data and the ranking of 12 candidate reference genes showed a slight difference. However, the ranking difference of reference genes between different algorithms was found by other authors (de Araujo et al. 2014; Cassol et al. 2016). Moreover, in our study, the most effective and stable candidate reference gene was assessed by R-package RankAggreg based on the ranking sequence of different algorithms. This evaluation improves the reliability of the most stable candidate reference genes.
The selection of the best stable candidate reference gene for the head and ovary of the queen was different under different reproductive status. The results suggest that the candidate reference genes have tissue-specific expression. This phenomenon was observed in the identification of reference genes for quantitative RT-PCR analysis of miRNAs in the Castor bean and bovine (Li et al. 2014; Cassol et al. 2016). To improve the accuracy of qPCR, multiple combinations of reference genes were recommended. The tool geNorm was developed to calculate the pairwise variation (Vn/n+1) between the normalization genes to determine the optimal number of reference genes. When the pairwise variation value was below 0.15, the addition of another reference gene was not required (Vandesompele et al. 2002). Based on the geNorm calculation, the combination of two candidate reference genes is sufficient to normalize the miRNA expression under different reproductive status (Fig. S2).
ACTB and GAPDH were commonly used as reference genes in bumblebee gene function analysis for their stable expression (Hornakova et al. 2010; Li et al. 2010; You et al. 2010). They exhibited substantial variation in the head and ovary of queens under different reproductive status, especially the poor expression stability of GAPDH in the ovary (Figure 1). Both genes were not recommended to be reference genes in miRNA quantification from our study results. Thus, we suggest that it is important to identify and validate a stable reference gene(s) for particular tissues and different biological status prior to publishing results of the miRNA expression based on the reference gene method.
The candidate reference miRNAs in this study were selected according to Sun’s result (Sun et al., 2015). It was reported that these high abundant miRNAs expressed stably in head and ovary under different reproductive statuses. Our results demonstrated that the high expression miR-277 was stable in the ovary between egg-laying and non-egg-laying queen. Interestingly, miR-277 were listed as the highly expressed microRNA in the brain and fat body of honey bees related to the expression level of vitellogenin (Nunes et al. 2013). Therefore, those high-expression miRNAs in the head and ovary of bumblebee may become the best candidate reference gene.
To the best of our knowledge, this report presents the first attempt to validate a set of candidate reference genes in the bumblebee queen head and ovary at different reproductive status. The transition of the queen’s reproductive status is an important physiological activity of bumblebee, which is a hot topic in recent years (Manfredini et al. 2017). Therefore, those reference genes evaluated in this study will be very useful for further miRNA expression analysis of head and ovary in the bumblebee queen, especially in the analysis of miRNAs related to oviposition.
This result suggests that the expression stability of endogenous reference gene is complex, and different biological samples should be tested for the stability of reference genes prior to use. Moreover, different algorithms should be applied to improve the reliability of the candidates. In addition, the commonly used reference genes (ATCB and GAPDH) were found to be unsuitable as candidates for the normalization of miRNA in the queen’s head and ovary under different reproductive status. Therefore, this study’s results indicate that the combination of miR-275 and miR-7 for the head and the combination of miR-277 and miR-317 for the ovary are the most reliable reference genes.
References
An, J., Huang, J., Shao, Y., Zhang, S. and Wang, B., et al. (2014) The bumblebees of North China (Apidae, Bombus Latreille). Zootaxa 3830, 1–89.
Andersen, C. L., Jensen, J. L. and Orntoft, T. F. (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64 (15), 5245–5250.
Baley, J. and Li, J. (2012) MicroRNAs and ovarian function. J. Ovarian Res. 5, 8.
Bartel, D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–33.
Cassol, D., Cruz, F. P., Espindola, K., Mangeon, A. and Müller, C., et al. (2016) Identification of reference genes for quantitative RT-PCR analysis of microRNAs and mRNAs in castor bean (Ricinus communis L.) under drought stress. Plant Physiol. Bioch. 106, 101–107.
de Araujo, M. A., Marques, T. E., Taniele-Silva, J., Souza, F. M. and de Andrade, T. G., et al. (2014) Identification of endogenous reference genes for the analysis of microRNA expression in the hippocampus of the pilocarpine-induced model of mesial temporal lobe epilepsy. PLoS One 9 (6), e100529.
Dheda, K., Huggett, J. F., Bustin, S. A., Johnson, M. A. and Rook, G. (2004) Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37, 112–114, 116, 118–119.
Guo, X., Su, S., Geir, S., Li, W. and Li, Z., et al. (2016) Differential expression of miRNAs related to caste differentiation in the honey bee, Apis mellifera. Apidologie 47 (4), 495–508.
Hornakova, D., Matouskova, P., Kindl, J., Valterova, I. and Pichova, I. (2010) Selection of reference genes for real-time polymerase chain reaction analysis in tissues from Bombus terrestris and Bombus lucorum of different ages. Anal. Biochem. 397 (1), 118–20.
Huggett, J., Dheda, K., Bustin, S. and Zumla, A. (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 6 (4), 279–84.
Kang, K., Peng, X., Luo, J. and Gou, D. (2012) Identification of circulating miRNA biomarkers based on global quantitative real-time PCR profiling. J. Anim. Sci. Biotechno. 3, 4.
King, I. N., Qian, L., Liang, J., Huang, Y. and Shieh, J. T. (2011) A genome-wide screen reveals a role for microRNA-1 in modulating cardiac cell polarity. Dev. Cell 20, 497–510.
Li, D., Liu, H., Li, Y., Yang, M. and Qu, C. (2014) Identification of suitable endogenous control genes for quantitative RT-PCR analysis of miRNA in bovine solid tissues. Mol. Biol. Rep. 41, 6475–6480.
Li, J., Huang, J., Cai, W., Zhao, Z. and Peng, W., et al. (2010) The vitellogenin of the bumblebee, Bombus hypocrita: studies on structural analysis of the cDNA and expression of the mRNA. J. Comp. Physiol. B. 180 (2), 161–170.
Li, L., Liu, F., Li, W., Li, Z. and Pan, J., et al. (2012) Differences in microRNAs and their expressions between foraging and dancing honey bees, Apis mellifera L. J. Insect Physiol. 58 (11), 1438–43.
Livak, K. J. and Schmittgen, T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25 (4), 402–8.
Macedo, L. M. F., Nunes, F. M. F., Freitas, F. C. P., Pires, C. V. and Tanaka, E. D., et al. (2016) MicroRNA signatures characterizing caste-independent ovarian activity in queen and worker honeybees (Apis mellifera L.). Insect Mol. Biol. 25 (3), 216–226.
Manfredini, F., Romero, A. E., Pedroso, I., Paccanaro, A. and Sumner, S., et al. (2017) Neurogenomic signatures of successes and failures in life-history transitions in a key insect pollinator. Genome Biol. Evol. 9 (11), 3059–3072.
Marco, A., Ninova, M. and Griffithsjones, S. (2013) Multiple products from microRNA transcripts. Biochem. Soc. T. 41, 850–854.
Nunes, F. M. F., Ihle, K. E., Mutti, N. S., Simões, Z. L. and Amdam, G. V. (2013) The gene vitellogenin affects microRNA regulation in honey bee (Apis mellifera) fat body and brain. J. Exp. Biol. 216 (Pt 19), 3724–3732.
Pfaffl, M. W., Tichopad, A., Prgomet, C. and Neuvians, T. P. (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26 (6), 509–15.
Pihur, V., Datta, S. and Datta, S. (2009) RankAggreg, an R package for weighted rank aggregation. BMC Bioinformatics 10, 62.
Plasterk, R. H. (2006) Micro RNAs in animal development. Cell 124 (5), 877–81.
Quah, S., Breuker, C. J. and Holland, P. W. (2015) A Diversity of Conserved and Novel Ovarian MicroRNAs in the Speckled Wood (Pararge aegeria). PLoS One 10 (11), e0142243.
Rozen, S. and Skaletsky, H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–86.
Serafin, A., Foco, L., Blankenburg, H., Picard, A. and Zanigni, S. (2014) Identification of a set of endogenous reference genes for miRNA expression studies in Parkinson’s disease blood samples. BMC Research Notes 7, 715.
Silver, N., Best, S., Jiang, J. and Thein, S. L. (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7, 33.
Sun, D., Dong, J., Huang, J., He, S. and Wu, J. (2015) Solexa sequencing and bioinformatics analysis of microRNA in the queen head of the bumblebee, Bombus lantschouensis. Acta Entomol Sin 58 (12), 1291–1299.
Tichopad, A., Didier, A. and Pfaffl, M. W. (2004) Inhibition of real-time RT–PCR quantification due to tissue-specific contaminants. Mol. Cell. Probe. 18, 45–50.
Vandesompele, J., Preter, K. D., Pattyn, F., Poppe, B. and Roy, N. V. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, 31–34.
Velthuis, H. H. W. and van Doorn, A. (2006) A century of advances in bumblebee domestication and the economic and environmental aspects of its commercialization for pollination. Apidologie 37 (4), 421–451.
Williams, P. H., Bystriakova, N., Huang, J., Miao, Z. and An, J. (2015) Bumblebees, climate and glaciers across the Tibetan plateau (Apidae: Bombus Latreille). Syst. Biodivers. 13 (2), 164–181.
Xie, F., Xiao, P., Chen, D., Xu, L. and Zhang, B. (2012) miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80 (1), 75–84.
Yeninar, H., Duchateau, M. J., Kaftanoglu, O. and Velthuis, H. (2000) Colony developmental patterns in different local populations of the Turkish bumble bee, Bombus terrestris dalmatinus. J. Apicult. Res. 39, 107–116.
You, H., Wan, H., Li, J. and Jin, B. R. (2010) Molecular cloning and characterization of a short peptidoglycan recognition protein (PGRP-S) with antibacterial activity from the bumblebee Bombus ignitus. Dev. Comp. Immunol. 34 (9), 977–985.
Zhou, Z., Zhang, H., Liang, C., Zou, Y. and Dong, J., et al. (2015) Foraging preference of the honeybee Apis mellifera and the bumblebee Bombus lantschouensis (Hymenoptera: Apidae) in peach greenhouse. Acta Entomol. Sin. 58, 1315–1321.
Acknowledgements
The authors would like to thank Prof. Jiandong An and Dr. Tolera Kumsa Gemeda for their invaluable comments.
Funding
This work was funded by China Agriculture Research System (CARS-44), the Agricultural Science and Technology Innovation Program (CAASASTIP-2018-IAR), and the Natural Science Foundation of China (U1603108).
Author information
Authors and Affiliations
Contributions
JD and JH conceived this research and designed experiments; JH, JL, and JW participated in the design and interpretation of the data; JD performed experiments and analysis; JD and JH wrote the paper and participated in the revisions of it. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Additional information
Manuscript editor: Marina Meixner
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Identification de gènes de référence appropriés pour la quantification des miARN dans la réponse à la reproduction des bourdons (Hymenoptera: Apidae)
bourdon / microARN / gène de référence / quantification en temps réel / normalisation
Identifizierung geeigneter Referenzgene für die Quantifizierung von miRNA in Abhängigkeit vom reproduktiven Status bei Hummeln (Hymenoptera: Apidae)
hummel / microRNAs / referenzgen / echtzeitquantifizierung / normalisierung
Electronic supplementary material
ESM 1.
(PDF 3483 kb)
Rights and permissions
About this article
Cite this article
Dong, J., Li, J., Huang, J. et al. Identification of suitable reference genes for miRNA quantitation in bumblebee (Hymenoptera: Apidae) response to reproduction. Apidologie 50, 40–50 (2019). https://doi.org/10.1007/s13592-018-0616-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13592-018-0616-9