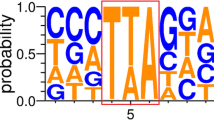
Abstract
Miniature inverted-repeat transposable elements (MITEs) are class II, non-autonomous DNA transposons that occupy a large portion of the genome, most in an inactive state. Because transposition of MITEs can have a broad impact on the structure and function of the genome, it is important to identify activated MITEs and analyze their propensity for transposition. However, to date the activity of only a few MITEs has been analyzed. In this study, MITE activation during the transformation processes in Chinese cabbage was analyzed by using next-generation sequencing. Using genome wide analysis, we found PTE-1 was activated during the transformation process. The active transposition of PTE-1 was analyzed by PCR amplification. We determined the sequence of PTE-1 by cloning the PCR products. Based on its target site duplications sequence and terminal inverted repeats structure, we inferred that the element belongs to the Tourist family. The characteristics of PTE-1, including structure and copy number, were identified by bioinformatics approaches. The results suggest that PTE-1 activation could be induced by the transformation process and reveal the first detection of activated MITE in tissue culture derived from Brassica rapa plants.






Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author upon request.
References
Benjak A, Boué S, Forneck A, Casacuberta JM (2009) Recent amplification and impact of MITEs on the genome of grapevine (Vitis vinifera L.). Genome Biol Evol 1:75–84
Bureau TE, Wessler SR (1992) Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell 4:1283–1294
Chen J, Hu Q, Zhang Y, Lu C, Kuang H (2013) P-MITE: a database for plant miniature inverted-repeat transposable elements. Nucleic Acids Res 42:D1176–D1181
Dai S, Hou J, Long Y, Wang J, Li C, Xiao Q, Jiang X, Zou X, Zou J, Meng J (2015) Widespread and evolutionary analysis of a MITE family Monkey King in Brassicaceae. BMC Plant Biol 15:149
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Feschotte C, Swamy L, Wessler SR (2003) Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with Stowaway miniature inverted repeat transposable elements (MITEs). Genetics 163:747–758
González J, Petrov D (2009) MITEs—the ultimate parasites. Science 325:1352–1353
Grandbastien MA, Spielmann A, Caboche M (1989) Tnt1, a mobile retroviral-like transposable element of tobacco isolated by plant cell genetics. Nature 337:376–380
Hirochika H (1997) Retrotransposons of rice: their regulation and use for genome analysis. Plant Mol Biol 35:231–240
Hirochika H, Sugimoto K, Otsuki Y, Kanda M (1996) Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci USA 93:7783–7787
Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, McCouch SR, Wessler SR (2003) An active DNA transposon family in rice. Nature 421:163–167
Jiang N, Feschotte C, Zhang X, Wessler SR (2004) Using rice to understand the origin and amplification of miniature inverted repeat transposable elements (MITEs). Curr Opin Plant Biol 7:115–119
Kaeppler SM, Kaeppler HF, Rhee Y (2000) Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol 43:179–188
Kikuchi K, Terauchi K, Wada M, Hirano H (2003) The plant MITE mPing is mobilized in anther culture. Nature 421:167–170
Lee M, Phillips RL (1988) The chromosomal basis of somaclonal variation. Annu Rev Plant Physiol Plant Mo Biol 39:413–437
Lisch D (2009) Epigenetic regulation of transposable elements in plants. Annu Rev Plant Biol 60:43–66
Makarevitch I, Waters AJ, West PT, Stitzer M, Hirsch CN, Ross-Ibarra J, Springer NM (2015) Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet 11:e1004915
Márton L, Hrouda M, Pécsváradi A, Czako M (1994) T-DNA-insert-independent mutations induced in transformed plant cells during Agrobacterium co-cultivation. Transgenic Res 3:317–325
McClintock B (1984) The significance of responses of the genome to challenge. Science 226:792–801
Park JH, Kim HS, Lee GH, Yu JG, Park YD (2016) Stable inheritance of an integrated transgene and its expression in phenylethylisothiocyanate-enriched transgenic Chinese cabbage. Korean J Hort Sci Technol 34:112–121
Peschke VM, Phillips RL (1991) Activation of the maize transposable element Suppressor-mutator (Spm) in tissue culture. Theor Appl Genet 81:90–97
Peschke VM, Phillips RL, Gengenbach BG (1987) Discovery of transposable element activity among progeny of tissue culture-derived maize plants. Science 238:804–807
Phillips RL, Kaeppler SM, Olhoft P (1994) Genetic instability of plant tissue cultures: breakdown of normal controls. Proc Natl Acad Sci 91:5222–5226
Sampath P, Murukarthick J, Izzah NK, Lee J, Choi H, Shirasawa K, Choi B, Liu S, Nou I, Yang TJ (2014) Genome-wide comparative analysis of 20 miniature inverted-repeat transposable element families in Brassica rapa and B. oleracea. PLoS ONE 9:e94499
Van Die IM, Bergmans HE, Hoekstra WP (1983) Transformation in Escherichia coli: studies on the role of the heat shock in induction of competence. Microbiology 129:663–670
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun J, Bancroft I, Cheng F, Huang S, Li X, Hua W, Wang J, Wang X, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Yang G, Lee Y, Jiang Y, Shi X, Kertbundit S, Hall TC (2005) A two-edged role for the transposable element Kiddo in the rice ubiquitin2 promoter. Plant Cell 17:1559–1568
Yang G, Nagel DH, Feschotte C, Hancock CN, Wessler SR (2009) Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. Science 325:1391–1394
Yu JG, Lee GH, Park YD (2016) Characterization of a drought-tolerance gene, BrDSR, in Chinese Cabbage. Korean J Hort Sci Technol 34:102–111
Acknowledgements
This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01365201)” Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Contributions
SA Kim performed the majority of the experiments and data analysis. YJ Jeon contributed to the activation experiment and data analysis. JS Park contributed to the development of the transgenic lines. YD Park designed the experiments and analyzed the data. SA Kim and YD Park wrote the manuscript. All authors contributed to and corrected the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Tae-Ho Han, Ph.D.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, Sa., Jeon, Y., Park, JS. et al. Discovery of PTE-1, Tourist-like miniature inverted repeat transposable element (MITE), and its activation in transgenic Brassica rapa ssp. pekinensis plants. Hortic. Environ. Biotechnol. 60, 955–965 (2019). https://doi.org/10.1007/s13580-019-00181-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13580-019-00181-1