Abstract
Graylings (Thymallus) are among the less well-studied groups of salmonid fishes, especially across their Asian distribution range. Here we perform a comprehensive global review of their phylogeography, systematic diversity and range distributions, including biogeographic reconstruction and assessment of both conservation and taxonomic status of each species. Based on a mitogenomic phylogenetic analysis, three approaches to the delineation of molecular operational units, and evaluation of 15 a-priori defined species, we provide biological support for the recognition of 13 grayling species, plus two additional species tentatively. Several instances of paraphyly and its potential effect on systematic inferences are discussed. Overall, the genus displays increasing species diversity and decreasing range size from higher to lower latitudes and ancestral trait reconstruction supports an East Asian origin for extant diversity, most likely centred in the Amur River drainage. Europe’s colonization by Thymallus took place as early as the late Miocene, at least two colonisations of North America are supported, and multiple dispersal events likely took place into Western Siberia. The conservation status for the 15 taxa was estimated to be: 6 least concern, 1 near-threatened, 2 vulnerable, 3 endangered and 3 data deficient.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salmonidae contains many of the most economically and ecologically important temperate freshwater fishes, which, due to their common tetraploid ancestry (Allendorf and Thorgaard 1984), attract considerable attention in evolutionary research. The subfamily Thymallinae (graylings), considered a distinct family by Skurikhina et al. (1985) and Osinov and Lebedev (2000), is less well known, although the recognized species diversity has increased considerably over the past 15 years (Antonov 2004; Knizhin et al. 2006a, 2007; Knizhin and Weiss 2009). Additionally, increased conservation concerns over European grayling Thymallus thymallus (Dawnay et al. 2011; Weiss et al. 2013; Mueller et al. 2018), as well as its recent whole-genome sequencing (Varadharajan et al. 2018; Sävilammi et al. 2019), have opened the doors to more focused evolutionary and conservation management research (Papakostas et al. 2014; Mäkinen et al. 2016; Huml et al. 2018). However, a pre-requisite to detailed evolutionary research and conservation is a clear understanding of species diversity, systematic relationships and distributions. The last comprehensive phylogenetic analysis of Thymallus (Froufe et al. 2005) became obsolete due to the description of a number of new species (e.g. T. ligericus) (Table 1). A recent annotated checklist of grayling species diversity noted that several species are of questionable status, or require additional research (Dyldin et al. 2017).
The majority of grayling species occur in Asia, occupying some of the world’s largest river drainages. Siberia’s complex paleo-hydrology has shaped the diversity and distribution of grayling throughout Eurasia (Koskinen et al. 2002; Froufe et al. 2003a, 2005). This is illustrated by the phylogeographic patterns in the Altai-Sayan mountain region (Knizhin and Weiss 2009; Weiss et al. 2020a) or the three sympatric species with allopatric origins found in the Bureya River, Amur River drainage (Weiss et al. 2020b). High phylogeographic complexity is also found in Europe within the Danube River drainage as well as the larger Ponto-Caspian basin (Weiss et al. 2002, 2013; Gum et al. 2009; Marić et al. 2012, 2014).
Regional phylogeographic studies have further contributed to understanding within-genus diversity. For instance, Knizhin et al. (2008) demonstrated that different grayling phenotypes in the endorheic basin of Western Mongolia can be phylogenetically assigned to T. brevirostris. Thymallus brevicephalus found in the upper Irtysh River drainage in Kazakhstan is the sister taxon to T. brevirostris (Weiss et al. 2020a). While such studies combined single-gene mtDNA analysis with population genetic and morphological analysis, they may lack phylogenetic resolution due to the limited length of the mtDNA sequence analysed.
Population genetic investigations of grayling across their range support that most, if not all species thus far described are either allopatric or when found in sympatry show little to no introgression (Koskinen et al. 2002; Froufe et al. 2003b; Weiss et al. 2006, 2007, 2020b). Thus, it is likely that mtDNA-based phylogenetic analyses of grayling reflect evolutionary relationships of the entire genome. Mitochondrial genomes (henceforth ‘mitogenomes’) present a significant improvement in resolution over analyses based on one or a few mtDNA genes (Miya and Nishida 2015; He et al. 2018; Boo and Hughey 2019). Low coverage genome-wide short-read sequencing can be used to recover mtDNA even from relatively degraded samples, due to the high mtDNA copy number per cell (Miller et al. 2012; Liedigk et al. 2015). Despite such advantages, to date, only a single application of mitogenomes to grayling phylogeny was carried out (Ma et al. 2016), whereby only eight mitogenomes were sequenced across seven species.
DNA-based species delineation methods have become standard tools for delineating molecular operational taxonomic units (MOTUs) (e.g. Lopes-Lima et al. 2019) or uncovering potentially cryptic species (e.g. Pan et al. 2019) in many organisms, including freshwater fishes (Patil et al. 2018; Berbel-Filho et al. 2018; Corral-Lou et al. 2019). The results of such methods are dependent on the choice of samples and loci (Ritchie et al. 2016; Sukumaran and Knowles 2017) and must be carefully interpreted. However, for a widespread genus like Thymallus, such analyses should allow a standardized evaluation of historically described taxa, which have received limited attention from modern molecular analyses.
In this context, a phylogenetic analysis of the genus Thymallus was carried out, using newly obtained mitogenomes via low-coverage whole-genome sequencing, together with already available mitogenome sequences. Using comprehensive taxon sampling and literature review, we aimed to: (a) produce an updated robust phylogeny of graylings across their global range; (b) evaluate current systematics using a set of species delineation methods; (c) provide an overview of the distribution and conservation status of each species; (d) reconstruct ancestral biogeographic patterns and (e) provide an up-to-date taxonomic overview. More broadly, we aim to provide a baseline for future research on evolutionary patterns, taxonomic diversity, conservation and biogeography of grayling throughout the world.
Methods
DNA extraction and sequencing
Whole genomic DNA of 47 grayling representing species across their global range was extracted from fin-clips stored in 96% ethanol using a high-salt protocol (Sambrook et al. 1989) (Table S1, Electronic Supplementary Material). Sample choice was influenced by the availability of published mitogenomes, aiming for a data set covering the global diversity of the genus. Included are most species listed in Dyldin et al. (2017) and the newly described T. ligericus (Persat et al. 2019) (Table 1). For each sample, Illumina paired-end libraries (TrueSeq DNA PCR-Free kit) were prepared and ~ 2 Gb of raw paired-end 150 bp (PE150) reads were sequenced on an Illumina Platform at Macrogen (Seoul, South Korea) and Novogene (UK, Europe).
Mitogenome assembly, annotation and characterization
Whole mitogenome assemblies were performed using a seed and an extend algorithm in NOVOplasty v2.7.2 (Dierckxsens et al. 2016), and the baiting and iterative approach in MITObim v1.9.1 (Hahn et al. 2013) following the authors’ guidelines and using default parameters. Additionally, six mitogenomes of T. thymallus and eight from T. ligericus were produced with the long-range PCR protocol described in Denys et al. (2020). The 61 new mitogenomes were annotated with MitoAnnotator (Iwasaki et al. 2013) via the web-interface (http://mitofish.aori.u-tokyo.ac.jp/, last accessed 28.06.2019). Annotations were converted to standard feature table format using a custom script (https://github.com/chrishah/MitoFish2tbl) and deposited to GenBank (MT062993-MT063053) and validated by comparison to published data (Table S1).
Phylogenetic analyses
The sequenced mitogenomes were aligned in MAFFT (Katoh and Standley 2013) together with mitogenomes retrieved from GenBank for 31 Thymallus spp. and three outgroup taxa (Table S1). For subsequent analyses, the mtDNA control region (CR) was removed due to several repeat motifs. The best partition scheme for each gene or group of genes was estimated using PartitionFinder2 v2.1.1 (Lanfear et al. 2012). Maximum likelihood analyses (ML) were performed with the program RAxML-HPC2 Workflow v8.2.10 (Stamatakis 2016), using the general time reversible substitution model, with a gamma parameter and proportion of invariable sites (GRT + I + G) and 1000 bootstrap replicates. Bayesian phylogenetic inference (BI) was carried out in MrBayes v3.2.6 (Ronquist and Huelsenbeck 2003), using the best substitution models previously obtained in PartitionFinder2. Two independent runs were carried out (107 generations, one tree sampled every 100 generations), with default chains. PartitionFinder2, MrBayes and RAxML were run in CIPRES Science Gateway (Miller et al. 2010). Tracer v1.7.1 (Rambaut et al. 2018) was used to assess the effective sample size values (ESS) and determine the appropriate burn-in.
Species delineation methods
Two distance-based and one tree-based method were applied to determine molecular operational taxonomic units (MOTUs): the cluster tool implemented in BOLD (Ratnasingham and Hebert 2007), the Automatic Barcode Gap Discovery (ABGD) (Puillandre et al. 2012) using both an initial ABGD(i) and a recursive ABGD(r) partition, and an improved multi-rate version (mPTP; Kapli et al. 2017) of the Poisson Tree Processes (Zhang et al. 2013). The BOLD method accepts COI sequences only, but ABGD and mPTP were applied to the mitogenome dataset. For BOLD, COI sequences were retrieved from aligned mitogenomes and uploaded on the BOLD platform. ABGD was run online (https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html; last accessed 20.02.2020), on the same dataset used for the phylogeny. The relative gap width was set to 1.0 and uncorrected p distances were used as a substitution model; remaining parameters were set as default. Finally, the mPTP method was applied to the Bayesian tree (without outgroups) using an online platform (https://mptp.h-its.org/#/tree; last accessed 23.01.2020). Samples were also assigned to species based on a combination of phenotypic identification, type locality, a-priori range knowledge and GenBank annotations. Uncorrected p distances were calculated among species or MOTUs for each delineation method (putative species, BOLD, ABGD (i), ABGD(r), mPTP using MEGA v10.0.5 (Kumar et al. 2018)) and frequency histograms of each were made using SPSS v26. For these calculations, four taxa listed in Table 1 were excluded due to the lack of evidence supporting their biological meaningfulness and/or our inability to assign a sample to them due to ambiguity concerning their type localities. Three of these taxa (T. mertensii, T. pallasii, T. signifer) are herein grouped within T. arcticus sensu lato (s.l.) and all are discussed in the results and discussion as well as Appendix 1 (Electronic Supplementary Materials). MOTU delineation was used as one line of potential evidence (sensu de Queiroz 2007) to support the existence of species. MOTU support by multiple algorithms together with population genetic and phylogeographic data, phenotypic distinction and ecological niche occupation were used to infer a taxon’s biological recognition. Lack of MOTU support was not necessarily used to argue against species recognition if multiple lines of other biological evidence supporting a species status exist.
Time-calibrated phylogeny
A time-calibrated phylogeny was produced using BEAST v1.10.4 (Drummond and Rambaut 2007; Suchard and Rambaut 2009) in CIPRES science gateway (Miller et al. 2010). Mitogenomes of eight individuals from Salmoninae and Coregoninae were added to the dataset as outgroups needed for the calibration scheme. The best partition scheme was selected as mentioned above. BEAST analysis was done with unlinked substitution and clock models, and a linked tree model, using uncorrelated relaxed molecular clock priors with log-normal distributions (Drummond et al. 2006). The birth-death speciation process (Gernhard 2008) was chosen as a tree prior, given the dataset comprised intra- and inter-specific relationships. One calibration point was set at 50 MY for the most recent common ancestor (MRCA) of Salmonidae, using the fossil †Eosalmo driftwoodensis Wilson, 1977 as a minimum time constraint for the family (Crête-Lafrenière et al. 2012; Lecaudey et al. 2018). Prior parameters were implemented following Lecaudey et al. (2018) (lognormal distribution, offset: 50, mean: 10, SD: 1). Another calibration point was set using the expansion of T. baicalensis into Lake Baikal (Koskinen et al. 2002), adjusted to 0.13 MY (Normal distribution, mean: 0.12, SD: 0.1), following the most recent dating of the ‘Angara breakout’ (Arzhannikov et al. 2018).
A third calibration point was set at 7.6 MY (stem dating; lognormal distribution; mean:7.6; SD:1.5) based on fossil remains of †Thymallus latisulcatus Rückert-Ülkümen & Kaya, 1993 found in Yalova (otolith used for species description) and Yalakdere, Turkey (Rückert-Ülkümen and Kaya 1993). The fossils were located in the strata corresponding to the Khersonian-Maeotian transition, whose date was recently revised to 7.6 MY (Lazarev et al. 2020; Palcu et al. 2019). Since this fossil may or may not correspond to a direct ancestor of the extant European lineages, the calibration was placed on the stem. The substitution rate prior for each partition was set with a normal distribution and 1%/MY (average 0.01 and 0.004 SD) following the mtDNA molecule calibration for salmonids (Smith 1992) and the reported lower bound of the T. baicalensis expansion (Koskinen et al. 2002). Additional Quaternary fossils of T. arcticus have been reported from North America (Cumbaa et al. 1981; McAllister and Harington 1969), but these were not included, since it was not possible to determine with certainty where these fossils would fall in the phylogeny. They could represent an ancestor of all North America populations, or only one of the lineages, or be placed within one of the lineages. Since the North American lineages are not monophyletic, using these fossils to delimit a minimum or maximum age for the split of both NA lineages would also require assuming where the split took place, and such an assumption might be less than informative. Analysis was performed with three 30 million MCMC iteration runs, sampling every 3000 runs. Burn-in and run convergence (ESS > 200) were determined using Tracer v1.7.1 (Rambaut et al. 2018). Independent runs were joined in LogCombiner v1.10.4 and the final tree produced in TreeAnnotator v1.10.4 (both in the BEAST package) and formatted in FigTree v1.4.4 (Rambaut 2012).
Species distributions and taxonomic review
Approximate species distributions were estimated based on both published and unpublished information (see Fig. 1). The area of occupancy (AOO) was calculated using ArcMap v10.7 (ESRI 2019) and hydrological network data from HydroAtlas v1.0 (Linke et al. 2019), as the total hydrological network area of lakes plus rivers larger than Strahler level three, a more realistic measure than a 2 × 2 km grid overlay (see Gomes-dos-Santos et al. 2019). Based on literature review, and present results, we comment on the biological status of each taxon, and review their taxonomic and conservation status based on IUCN criteria.
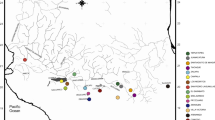
Map showing the global distribution range of Thymallus species. Information on sampling sites and species is given in Table S1. Numbers in the map refer to known contact zones of the following species: 1 = T. arcticus s.l. and T. baicalensis in the lower Enisei River; 2 = T. arcticus s.l. and T. baicalolenensis in the lower Lena River; 3 = T. nikolskyi and T. baicalensis in tributaries of the upper Ob River; 4 = T. baicalolenensis and T. baicalensis in tributaries of Lake Baikal; 5 = T. grubii, T. tugarinae and T. baicalolenensis (not all found in contact throughout this zone) in the upper and middle Amur drainage (including its tributaries such as the Ingoda, Argun and Zeya rivers); 6 = T. flavomaculatus, T. tugarinae and T. baicalolenensis in the Uda River drainage, 7 = T. burejensis, T. grubii and T. baicalolenensis in the upper Bureya River; 8 = T. tugarinae and T. flavomaculatus in tributaries of the lower Amur River
Historical biogeography and ancestral area reconstruction
To reconstruct the ancestral ranges of extant Thymallus diversity, we performed a Bayesian phylogeographic inference in discrete space using the Bayesian Stochastic Search Variable Selection (BSSVS; Lemey et al. 2009) implemented in BEAST. Considering both the current and paleo-hydrological drainage patterns, we considered seven biogeographic regions: Europe (west of Urals), Western Siberia (Kara Sea basin); East Siberia (north-flowing rivers, and Bering Sea); East Asia excluding the Amur (Okhotsk, Japan, Yellow and East China sea drainages); Amur River drainage; North America and the endorheic drainages of Western Mongolia. We used the same dataset, models and prior settings as in the dating analysis. BSSVS parameters were as follows: symmetric substitution model, strict location-trait clock and an exponential distribution for the location rates prior (mean = 1, offset = 0).
Results
Mitogenome analyses
Low-coverage genome sequencing recovered mitogenomes for all 47 samples analysed. With on average 18 million genomic reads per sample, the number of mitochondrial reads ranged from almost 3 to more than 50 thousand per sample for an organelle coverage ranging from 25x to 390x (Table S2). Reads and circularized molecules were deposited on GenBank (MT062993-MT063053, BioProject: PRJNA604892) (Table S2). The mitogenomes included 13 protein-coding genes, 22 tRNAs, 2 rRNAs and the CR (see Fig. S1, Electronic Supplementary Material). Average nucleotide composition and gene order were consistent across all individuals (Fig. S1).
Phylogenetic analyses and species delineation
ML and BI analyses showed the same topology; thus, only the Bayesian phylogenetic tree is represented (Fig. 2). The tree depicts three main groups of species, with the first split separating East Asian species (excluding T. burejensis) from all others, and the next split separates the three accepted European species from those in Siberia and the Altai, including T. burejensis. Pairwise divergence among a priori defined species ranged from 0.3% for T. nigrescens and T. baicalensis, to 5.5% between T. tugarinae and T. aeliani (Table S3). Net uncorrected p distances among all MOTUs revealed a bi-modal distribution, with a minor mode at 0.1–0.2% and a major mode centred on approximately 5% sequence divergence (Fig. S2). Congruence between well-supported clades, all delineation algorithms and a priori species assignment occurred for T. baicalolenensis, T. burejensis, T. aeliani and T. ligericus. Thymallus baicalensis was not monophyletic due to the placement of T. nigrescens within T. baicalensis. Thymallus tugarinae was also not completely congruent due to a divergent haplotype from the Kievka River, which drains to the Sea of Japan (KY078218).
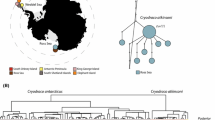
Bayesian phylogenetic reconstruction based on whole mitogenome dataset (excluding the CR). Branch support values represent Bayesian posterior probabilities (above nodes) and maximum likelihood values (below nodes). An asterisk symbolizes that both values were ≥ 90%. To the right of the tree shown in grey bars are the results of three MOTU delineation methods. Each bar represents one MOTU: 12 for BOLD, 16 for mPTP and 21 for ABGDa (indicates both initial and recursive partition). Taxa with a smaller font positioned right of the MOTU bars represent names in use for which there is no biological support and thus may represent junior synonyms. Species are coloured according to Fig. 1
The most conservative delineation algorithm was BOLD, recognizing 12 MOTUs. The two ABGD schemes recognized 21 MOTUs, and the mPTP recognized 16 MOTUs. Paraphyletic relationships were inferred for T. thymallus and T. flavomaculatus. The former paraphyly results from a well-supported clade representing Alpine haplotypes from the upper Danube (Weiss et al. 2002, 2013) resolved as the sister clade (1.5% divergent) of T. ligericus and delineated as one (BOLD) or two (mPTP and ABGD) MOTUs. The node containing T. grubii, T. flavomaculatus and T. yaluensis represents the common ancestor of T. flavomaculatus. The earliest split and most divergent clade within T. arcticus s.l., delineated as a MOTU by mPTP and ABGD, was represented by haplotypes (MT063015-017) from north-eastern British Columbia in Canada (Table S1). No additional Arctic taxa (e.g. T. pallasii, T. mertensii or T. signifer) (see Appendix 1) could be recognized by MOTU delineation, though, as noted above, there is ambiguity concerning the assignment of these taxa to a location or sample in our data set. Phylogenetic analysis resolved the sister species T. brevirostris and T. brevicephalus (0.4% divergent), but not MOTU delineation. Similarly, phylogenetic analysis resolved the sister species T. nikolskyi and T. svetovidovi (0.6% divergent), but only the mPTP algorithm delineated them as distinct MOTUs.
Time calibration and ancestral range reconstruction
The MRCA of Salmonidae was dated at 59.3 MY, at the end of Palaeocene (Fig. 3) while that for Thymallus was 9.11 MY, at the end of Miocene. The MRCA of three East Asian species, T. tugarinae, T. flavomaculatus and T. grubii, was estimated at 8.42 MY also in the late Miocene, similar to the split between European species and all others (7.54 MY), whereas the MRCA of all European species was estimated at 3.86 MY corresponding to the mid-Pliocene. The next oldest node (6.35 MY) represented the split between T. burejensis and remaining species. The estimated MRCA of T. arcticus s.l. and T. baicalolenensis 5.01 MY and that of all T. arcticus was 2.39 MY, in the Pliocene and early Pleistocene, respectively. The splits between two pairs of allopatric species in the Altai-Sayan mountain region [T. brevirostris and T. brevicephalus (0.85 MY), and T. svetovidovi and T. nikolskyi (0.77 MY)] were both dated to the mid-Pleistocene. The Amur River drainage was the most likely ancestral location of origin of the extant grayling diversity, with much higher support than every other option (Fig. 3). A single colonization of Europe from Western Siberia was predicted, but multiple exchanges may have occurred among the other considered regions, exemplified by the three-basin distribution of the young T. baicalolenensis, or two predicted ancestors of North American lineages over 1 MY apart.
Time-calibrated phylogeny and biogeographic analysis of Thymallus spp. inferred from the whole mitogenome (excluding the CR). Three fossil calibration points (panel a) and a biogeography-based substitution rate were used (details in text). Ancestral ranges were reconstructed as a discrete trait following the phylogeographic diffusion BSSVS method; nodes and branches are colour-coded according to region. Hypothesised dispersal events are shown in panel b. Potentially relevant paleo-climatic events such as the Maximum Ponto-Caspian extent (MPC), the following Fluvial Plains Period and the Eurasian Glacial Cycles are represented at the bottom. MSC, Messinian salinity crisis; KMT, Khersonian-Meotian transition
Discussion
Species delineation
Our results strongly support the biological validity of at least 13 grayling species globally, plus two additional species tentatively all but one supported by at least one MOTU delineation approach, the exception being T. nigrescens. Microsatellites and single mtDNA sequences allow no distinction between T. nigrescens and T. baicalensis (Koskinen et al. 2002; Kaus et al. 2019). Next-generation sequencing RAD data supported a 15–30 thousand year divergence between the two taxa (Roman et al. 2018). The taxon is morphologically distinct from T. baicalensis based on a small dorsal fin and a higher number of gill rakers (Knizhin and Weiss 2009; Olson et al. 2019), and thus, its occupation of a distinct ecological niche supports its status as a distinct species despite relatively low genetic divergence.
Mean pairwise distances among species (3.7%) conformed to minimum thresholds applied in other studies. The majority (> 90%) of vertebrate species differ from their nearest neighbour by > 2% (Avise et al. 1999) or > 3% (Hebert et al. 2004) using standard mtDNA gene markers. Such thresholds have been noted in many data sets involving fishes (e.g. Ward 2009; April et al. 2013). Our divergence values may be comparatively low as we use uncorrected p distances instead of model-corrected distances (see Srivathsan and Meier 2012), salmonid mtDNA exhibits a relatively slow substitution rate (ca. 1%/MY) (Smith 1992; Froufe et al. 2005) and our calculations include non-protein regions of the mitogenome (excl. the CR). Higher values are obtained when using protein-coding genes alone (Fig. S3). Besides the baicalensis/nigrescens species pair, two other pairwise distances in our data under these thresholds are between sister species pairs in the Altai-Sayan mountain region (T. brevirostris and T. brevicephalus; and T. nikolskyi and T. svetovidovi). These species pairs span allopatric drainages subject to catastrophic paleo-hydrological flooding (Weiss et al. 2020a), which may have played a role in their cross-basin colonization history.
Five additional valid Thymallus names (T. yaluensis, T. brevipinnis, T. signifer, T. pallasii and T. mertensii) are supported neither by MOTU delineation nor with existing morphological or genetic data (Fig. 2) (see Appendix I). Two phylogenetically well-supported clades, also supported by one or more delineation approaches, have no species assignment. The first of these makes T. thymallus paraphyletic and consists of upper Danubian haplotypes, forming a sister clade to T. ligericus. This relationship was not recovered in previous phylogenetic analyses based on mitochondrial CR (Weiss et al. 2002, 2013; Marić et al. 2014), underscoring the higher resolution of the whole mitogenome and/or the importance of balanced taxon sampling, but also the need for additional investigation (see Appendix I for additional discussion). The second of these unassigned clades involves haplotypes (MT063015–017) of T. arcticus s.l., representing a lineage that is believed to derive from the Nahanni glacial refuge (Stamford and Taylor 2004). This mtDNA lineage was among three lineages described in North America (Stamford and Taylor 2004), whereby two colonization events into North America were proposed. The possibility that these haplotypes could represent a distinct taxon must be better explored with population genetic analysis.
The paraphyly of the T. flavomaculatus clade in our tree may be based on the complex phylogeographic structure of both T. flavomaculatus and T. grubii combined with our limited sampling. Both T. flavomaculatus and T. tugarinae are found in tributaries of the Amur River drainage, but also coastal drainages flowing directly into the seas of Japan and Okhotsk (Antonov and Knizhin 2014) promoting fragmentation and divergence (Appendix I).
Biogeography
Species diversity and range size in Thymallus spp. reveal a latitudinal gradient with few species above the Arctic Circle and increasing diversity (Spearman’s rank − 0.819, P < 0.05) and decreasing range size (Spearman’s rank 0.785, P < 0.001) with decreasing latitude (Fig. 1). Ancestral trait reconstruction supports the present Amur River drainage as the place of origin for extant diversity (Fig. 3), with lineage sharing among coastal draining rivers reflecting drainage re-arrangements and dynamics of the Pleistocene (Grosswald 2009; Antonov 2012; Antonov and Mikheev 2016). Europe’s colonization by Thymallus took place somewhere in the Pliocene, between the split from extant Asian lineages in the late Miocene, and the European lineage crown age in the mid-Pliocene. We note that the rough estimates of the timing of these events are affected by the fossil calibration point placed on the stem leading to the European radiation of the genus, derived from the 7.6 MY-old fossil remains of †T. latisulcatus. A previous analysis lacking this calibration point resulted in ages about 20% younger, but still falling in the same epochs described here, and provided even higher support for an Amur ancestral range (all nodes > 80%). Higher connectivity among Arctic Ocean drainages is illustrated by at least two colonisations of North America from Eastern Siberia and is analogous to Holarctic or near Holarctic distributions in Pungitius (Guo et al. 2019), Esox (Skog et al. 2014), and Lota (Van Houdt et al. 2005). Dispersal into Western Siberian rivers seems to have taken place multiple times, most recently from Eastern Siberia and the Amur, after a much more ancient event. The suggested colonization of the endorheic basin in Mongolia from East Siberian basins evokes the still unconfirmed proposals of a Gobi-Amuran proto-glacial drainage system (Grosswald 1998). Coastal rivers of East Asia retained connectivity with the Amur during the Pleistocene, reflected by the absence of a geographically coherent monophyletic group. However, a Miocene-Pleistocene split and posterior isolation between the extant diversity in both basins cannot be excluded, due to the long un-split branches from Miocene to Pleistocene (Fig. 3). Overall, the biogeographic patterns of Thymallus are concordant with scenarios proposed for several taxa. Cobitid fishes reveal an early radiation in eastern Asia, and colonization of Europe from East Asia in the late Miocene (Šlechtová et al. 2008; Perdices et al. 2016). A colonization from northeast Asia to Europe is proposed for Carassius (Rylková et al. 2013), and similar colonization of Rhodeus (via vicariance) is inferred for the Pliocene (Bohlen et al. 2006), supported by the disjoint distribution and hypothesised vicariance of Margaritifera (Bolotov et al. 2016) a parasitic bivalve of Rhodeus. These studies, however, do not provide explicit paleo-hydrological pathways or events that may have facilitated these patterns. While some of these hypothesised colonization events are pre-Pleistocene, our knowledge at least of Pleistocene paleo-dynamics across Russia reveals numerous events of paleo-hydrological connectivity (via large paleo-lakes and mega floods) across Siberia and through the Ponto-Caspian region into Europe (Komatsu et al. 2016). These events have already served hypotheses concerning the phylogeographic structure of Thymallus, Hucho and Brachymystax across their Siberian range (Koskinen et al. 2002; Froufe et al. 2003a; Weiss et al. 2020a). Nonetheless, much work remains in matching the timing of specific events of dispersal and vicariance with specific paleo-events promoting cross-drainage connectivity and isolation.
The following section provides a very brief overview of the 15 focal taxa of our phylogenetic and MOTU analyses. Included is a general description of their geographic range and phylogenetic position as well as our opinion on each taxon’s validity as a species (i.e. regarding its biology) and its suggested conservation status based on IUCN criteria. For more detailed comments on these taxa and those for which we do not currently find biological support, or cannot assign to a sample, see Appendix I.
Amur and east Asian taxa
Thymallus tugarinae— Lower Amur grayling
Occurs in the middle to lower Amur River drainage and several coastal draining rivers including northwestern Sakhalin Island (Fig. 1). Highly divergent from its sister clade containing T. grubii (4.1%) and T. flavomaculatus (4.2%). Easily distinguished from sympatric T. flavomaculatus using meristic or morphological characters (Knizhin et al. 2007); reciprocally monophyletic with other Amur graylings (Fig. 2) (Froufe et al. 2003b, 2005); reproductive isolation supported (Froufe et al. 2003b). Shows significant phylogeographic structure across its range (Froufe et al. 2003b).
Taxonomic validity
A species reproductively isolated and easily distinguished from the sympatric T. flavomaculatus (see T. flavomaculatus below).
Conservation remarks
Area of occupancy exceeds 2000 sq. km. Suggested Global Status: Least Concern.
Thymallus flavomaculatus— Yellow-spotted grayling
Occurs in some coastal rivers draining into the seas of Japan and Okhotsk, as well as some lower Amur River tributaries (Fig. 1); overlaps considerably with T. tugarinae (Froufe et al. 2003b; Antonov and Knizhin 2011). Easily distinguished from T. grubii and sympatric T. tugarinae based on a characteristic yellow-orange spot located in the posterior area of the dorsal fin (Knizhin et al. 2006a).
Taxonomic validity
The taxon was originally described as a subspecies of T. grubii, widely distributed in the Amur River drainage. Whether or not the taxon is treated as a species or a sub-species is beyond the scope of this manuscript. In our analysis, T. flavomaculatus is paraphyletic (see additional comments below for T. grubii).
Conservation remarks
Coastal populations are more threatened by anthropogenic changes and overfishing than interior populations. Some range fragmentation is present and the area of occupancy may be as little as 400 sq. km. Suggested Global Status: Near threatened.
Thymallus grubii— Amur grayling
A small-sized grayling, easily diagnosed based on body and dorsal-fin colouration; occurs throughout the middle to upper Amur River drainage (Fig. 1). Significant phylogeographic structure is reported (Froufe et al. 2003b; Knizhin et al. 2004; Weiss et al. 2020b). Occurs in sympatry with T. burejensis and T. baicalolenensis in the upper Bureya River, where reproductive isolation is strong but not complete (Weiss et al. 2020b), and in sympatry with T. tugarinae in the lower Zeya River and Ingoda River, and with T. baicalolenensis in the upper Zeya River and upper Ingoda River (Antonov and Mikheev 2016).
Taxonomic validity
A species diagnosable from all other grayling (Weiss et al. 2020b). Could be also treated as the nominal species of a three-taxon aggregate, consisting of T. (grubii) grubii, T. (grubii) flavomaculatus and T. (grubii) yaluensis (see Article 6.2 in the International Commission on Zoological Nomenclature 1999) (see also Appendix I).
Conservation remarks
Occupies a relatively large range and several relatively pristine river systems. Suggested Global Status: Least Concern.
Thymallus burejensis— Bureya grayling
A robust-bodied grayling, endemic to the middle and upper reaches of the Bureya River. Occurs in sympatry in the upper Bureya River with T. baicalolenensis and T. grubii, and shows relatively strong (albeit not complete) reproductive isolation (Weiss et al. 2020b). Mitogenomic distances range from 3.0 to over 4.9% between T. burejensis and all other congeners.
Taxonomic validity
A species displaying significant reproductive isolation with two other grayling taxa.
Conservation remarks
The Bureya River is over 700 km in length but the mid-to-lower reaches have been heavily impacted by hydropower, eliminating or fragmenting portions of the specie’s range. Currently, its area of occupation does not exceed 100 sq. km. Suggested Global Status: Endangered.
European taxa
Thymallus aeliani —Adriatic grayling
Occupies the middle to upper reaches of the Soca River in Slovenia and tributaries of the Po and Adige rivers in Italy (Fig. 1). Divergent from T. thymallus (2.7%) and T. ligericus (3.6%) within the clade of European grayling taxa (Fig. 2). Meraner et al. (2014) reported significant regional structure in the Adige River drainage.
Taxonomic validity
A species based on its deep divergence to all other grayling and allopatric distribution in Adriatic draining rivers.
Conservation remarks
River engineering measures, hydropower expansion and water pollution are among widespread threats that have reduced at least 50% of the species range; introgression with non-native lineages is a major threat to T. aeliani and there are few pure genetic populations left (Sušnik et al. 2004; Meraner et al. 2014). The area of aquatic occupancy may be as little as 100 sq. km. Suggested global status: Endangered.
Thymallus thymallus—European grayling
Widely distributed (Fig. 1); until recently included all European stocks of grayling. They, along with T. aeliani and T. ligericus, are the only Thymallus species with a sub-terminal mouth. Significant phylogeographic structure throughout Western Europe (Weiss et al. 2002; Gum et al. 2009), and from the western Balkans and Caspian Sea catchment (Marić et al. 2012, 2014). The taxon is paraphyletic due to the systematic relationship to both T. ligericus and its sister clade of upper Danubian haplotypes.
Taxonomic validity
A species distinguished from all Asian grayling by a subterminal mouth and strict long-time allopatry to T. ligericus and T. aeliani.
Conservation remarks
Currently listed as a species of Least concern. Locally, and especially in the southern portions of its range, population declines or extinctions are widespread (see Weiss et al. 2013), leading to several endangered assignments at national levels. Suggested Global Status: Least Concern.
Thymallus ligericus— Loire grayling
Recently described endemic of the upper Loire River drainage in France (Persat et al. 2019) (Fig. 1). Populations remain genetically pure despite 50 years of stocking with foreign strains (Persat et al. 2016), suggesting they either outcompete foreign lineages (i.e. T. thymallus) or display reproductive isolation. Morphologically distinguished from T. thymallus by a more pointed snout, more inferior mouth and profuse spotting (Persat et al. 2019). In our analysis, they appear as a shallow, monophyletic clade, 1.5% divergent from T. thymallus haplotypes from the upper Danube drainage and 2.2% divergent from all T. thymallus samples. The zoogeographic origins of this species in the Loire basin are unknown.
Taxonomic validity
A species based on its morphological and genetic distinction and long-term isolation (2 MY or more) from grayling of adjacent river drainages (Rhine and Rhône) (Persat et al. 2016).
Conservation remarks
Populations are in decline and there are concerns of decreasing water flows and rising water temperatures. Its area of occupancy may not exceed 35 sq. km, but at least six or more fragmented populations exist. Suggested Global Status: Vulnerable.
Altai, Siberian and North American taxa
Thymallus arcticus s.l.—Arctic grayling
Occurs from just east of the Urals in Russia to Hudson Bay, Canada; a disjunct population in the Big Hole and Red Rock river drainages in Montana, USA (Fig. 1). In our analysis, T. arcticus s.l. is 2.6% divergent from its sister taxon T. baicalolenensis. Phylogeographic structure across the Arctic is weak; for example, the haplotype MT063012 near the type locality (Sob River, Ob) groups closely with haplotypes from the Lena River drainage and the Okhotsk Sea catchment in far eastern Russia. Haplotype MT063010 from the presumed type locality of T. pallasii (see Dyldin et al. 2017) (Appendix I) in eastern Siberia is intermediate between most North American haplotypes and those from Kamchatka, which some authors assign to T. mertensii.
Taxonomic validity
A species based on its clear genetic divergence to other taxa, morphological distinctiveness especially in the dorsal-fin size and colouration (albeit with regional variation), and confirmed reproductive isolation to T. baicalensis (Weiss et al. 2007) and T. baicalolenensis (Weiss et al. 2006). See comments in Appendix I concerning potential recognition of additional taxa, herein treated as T. arcticus s.l.
Conservation remarks
The global population of T. arcticus s.l. is listed by the IUCN as a species of Least concern (LC). Numerous reports exist of population size declines for the species locally, both in North America and Russia. Suggested Global Status: Least Concern.
Thymallus baicalensis —Baikal black grayling
Occurs throughout the Enisei River drainage including Lake Baikal and its major tributary the Selenga River (Fig. 1); also in some right-hand tributaries of the Ob River drainage (Mrassu and Kabyrza rivers) represented by haplotypes MT063026 and MT063025 (Fig. 2). Displays a net divergence between 1.6 and 1.9% to four species comprising its sister clade (T. nikolskyi, T. svetovidovi, T. brevicephalus and T. brevirostris). Based on Koskinen et al. (2002), multiple samples throughout Lake Baikal were in Hardy-Weinberg Equilibrium reflecting a single taxon occupying Lake Baikal; this inference is also supported by morphological and genetic data from Knizhin et al. (2006b). All species delineation approaches allocated both T. baicalensis and T. nigrescens as a single MOTU.
Taxonomic validity
A species based on clear genetic divergence to other taxa, distinct dorsal-fin colouration and multiple contact zones with little to no gene flow with other species (T. arcticus, T. baicalolenensis) (see Knizhin et al. 2006b; Weiss et al. 2007). Thymallus brevipinnis is suggested to be a synonym of T. baicalensis (Appendix I).
Conservation remarks
Local population declines and extinctions reported; threats include hydropower development, overfishing and pollution. However, the species has a very large distribution range and occupies many habitats that are in pristine or near-pristine condition. Suggested Global Status: Least Concern.
Thymallus baicalolenensis —Baikal-Lena grayling
Small-bodied grayling occurring throughout the Lena River drainage (Fig. 1). Also occurs in Lake Baikal tributaries, most notably the Barguzin River drainage, the Tiya River and Yakchinskie Lakes of the upper Angara River (Knizhin et al. 2006c, 2008; Kirillov and Knizhin 2014). Also found in the Uda River drainage, Sea of Okhotsk (Antonov and Knizhin 2011) and upper Amur River drainage (Antonov and Knizhin 2011; Antonov and Mikheev 2016), including the upper Bureya River, together with T. burejensis and T. grubii, where it could be diagnosed with 100% accuracy based on morphological characters (Weiss et al. 2020b). Its morphological and genetic distinction from T. arcticus s.l. is shown in Weiss et al. (2006) (therein referred to as T. a. lenensis) and Koskinen et al. (2002) (therein referred to as T. arcticus, Lena basin). Both its body and dorsal-fin colourations are highly distinct (Knizhin et al. 2008; Knizhin and Weiss 2009) from T. arcticus s.l. as well as all other members of the genus (Dyldin et al. 2017). Thymallus baicalolenensis is reciprocally monophyletic to T. arcticus s.l. with a net divergence of 2.6% (Fig. 2).
Taxonomic validity
A species showing relatively strong reproductive isolation to four species to which it comes into contact; T. baicalensis, T. arcticus s.l., T. burejensis and T. grubii.
Conservation remarks
Very large distribution area, occupies both large rivers and headwaters including small lakes. Found in numerous relatively remote and/or pristine systems. Suggested Global Status: Least Concern.
Thymallus brevicephalus —Shorthead or Markakol grayling
Reported endemic to Lake Markakol (Dyldin et al. 2017); probably not limited to this lake. Haplotypes LC168675 and MT063035 (Fig. 2) stem from samples in the upper Irtysh River drainage; population genetic analysis shows close affinity with samples from the Kara-Kaba River (Weiss et al. 2020a) (Fig. 1). Both our mitogenome analysis and a population genetic analysis in Weiss et al. (2020a) show a very close (0.4%) sister clade relationship to T. brevirostris (Fig. 2). More data concerning morphology and ecology is needed.
Taxonomic validity
Although closely related genetically to T. brevirostris, viewed as species based on highly distinct morphology (short jaws, no dentation) and ecology (predominantly benthivore), as well as strict allopatric occurrence to T. brevirostris.
Conservation remarks
Populations within Lake Markakol are in serious decline due to overfishing (perhaps > 50% across recent decades, M. Baimukanov, pers. comm.); listed as endangered in Kazakhstan. Its strict area of aquatic occupancy (Markakol Lake) is < 700 sq. km, but its distribution is likely considerably larger. Suggested Global Status: Data deficient.
Thymallus brevirostris— Mongolian grayling
Distributed across the large endorheic basin of Western Mongolia, extending into Tuva Republic, Russia (Fig. 1). Considered a large-growing, piscivorous grayling, with pronounced dentation on both jaws and vomer (Knizhin et al. 2008). Displays considerable phenotypic variability concerning head and jaw size and dentation (Knizhin et al. 2008; Weiss et al. 2020a). Displays a close (0.4%) sister clade relationship to T. brevicephalus from Lake Markakol (Fig. 2).
Taxonomic validity
Although closely related to T. brevicephalus, viewed as a species based on highly distinct morphology (large jaws, significant dentation), ecology (predominantly piscivorous) and strict allopatric occurrence to T. brevicephalus (see also Appendix I).
Conservation remarks
Overfishing (primarily illegal), hydropower development and spawning ground deterioration are a major concern. Suggested Global Status: Vulnerable.
Thymallus nigrescens— Chovsgul grayling
Endemic to Lake Chovsgul in Mongolia. The single mitogenome (MT063028) in our analysis is 0.3% divergent from T. baicalensis (Fig. 2). Traditional genetic markers (microsatellites and single mtDNA sequences) allow no distinction between T. nigrescens and T. baicalensis (Koskinen et al. 2002; Kaus et al. 2019).
Taxonomic validity
Treated as a species despite its very close relationship to T. baicalensis. Morphologically distinct from T. baicalensis based on a small dorsal fin and a high number of gill rakers (Knizhin et al. 2008; Olson et al. 2019), occupies a distinct allopatric ecological niche.
Conservation remarks
Listed as endangered in the Mongolia Red List (Ocock et al. 2006). Its habitat comprises the 2760 sq. km Lake Chovsgul. Substantial illegal harvest via gillnetting in the littoral zone has led to dramatic declines in population sizes (Free et al. 2015). Suggested Global Status: Vulnerable.
Thymallus nikolskyi —Upper Ob grayling
Originally reported from the upper Ob River drainage; exact distribution is unclear. May occur together with T. baicalensis in the Mrassu and Kabyrza rivers of the Ob River drainage (Fig. 1). Our analysis reveals a close (0.6%) sister relationship to T. svetovidovi from the upper Enisei River drainage, and considerably more divergence from the two other taxa, T. arcticus s.l. and T. brevicephalus, in the Ob River drainage (3.0% and 1.7%, respectively). Population genetic analysis shows no gene flow between T. nikolskyi and T. brevicephalus (Weiss et al. 2020a).
Taxonomic validity
Treated tentatively as a species whereby the distributions and genetic relationships of all grayling in the Altai-Sayan mountain region require further investigation (Weiss et al. 2020a).
Conservation remarks
Insufficient data on its diagnosis and distribution. Suggested Global Status: Data deficient.
Thymallus svetovidovi— Upper Enisei grayling
Recently described from the Sharga Gol River in Mongolia (Knizhin and Weiss 2009); occurs in headwater reaches of the Enisei River in Mongolia and possibly Tuva Republic (Fig. 1). Bright yellow caudal peduncle and fin is highly characteristic; a close (0.6%) sister taxon to T. nikolskyi from the upper Ob River drainage.
Taxonomic validity
A species based on a unique phenotype and genetic divergence from T. baicalensis. See Appendix I for comments on potential synonymies.
Conservation remarks
Known distribution range is limited (but uncertain), populations are reportedly dense and the river systems where this taxon is found are pristine. Thus, there are currently no threats to this taxon. Suggested Global Status: Data deficient.
Data availability
Data generated or analysed during this study consist of mitogenome sequences. These sequences are deposited in GenBank under Bioproject number PRJNA604892; all GenBank accession numbers used in our analysis are also listed invidivually in the Electronic Supplementary File (Table S1).
References
Allendorf, F. W., & Thorgaard, G. H. (1984). Tetraploidy and the evolution of salmonid fishes. In Evolutionary genetics of fishes (pp. 1–53). US: Springer. https://doi.org/10.1007/978-1-4684-4652-4_1.
Antonov, A. L. (2004). A new species of grayling Thymallus burejensis sp. nov. (Thymallidae) from the Amur Basin. Journal of Ichthyology, 44, 401–411.
Antonov, A. L. (2012). Influence of hydrological network reconstruction on formation of habitats for some fish species in the Amur River basin and adjacent territories. Izvestia Russian Geographical Society, 144(6), 30–37. [In Russian].
Antonov, A. L., & Knizhin, I.B. (2011). Distribution, ecological features, and possible trends in development of habitats of graylings (Thymallidae) in the Amur basin. Herald North-Eastern Scientific Center of the Far Eastern Branch of the Russian Academcy of Sciences 1, 41–48. [In Russian].
Antonov, A. L., & Knizhin, I.B. (2014). Graylings (Thymallidae) form the Amur basin: history of studies and modern concepts about diversity. Amur Zoological Journal, 6(3), 298–307. [In Russian].
Antonov, A. L., & Mikheev, I. E. (2016). New data about distribution of lower Amur Grayling Thymallus tugarinae and Baikal-Lena Grayling Thymallus sp. (Thymallidae) in the Basins of the Upper Amur River and Zeya River. Journal of Ichthyology, 56(2), 208–216. https://doi.org/10.1134/S0032945216020016.
April, J., Hanner, R. H., Mayden, R. L., & Bernatchez, L. (2013). Metabolic rate and climatic fluctuations shape continental wide pattern of genetic divergence and biodiversity in fishes. PLoS ONE, 8(7), e70296. https://doi.org/10.1371/journal.pone.0070296.
Arzhannikov, S. G., Ivanov, A. V., Arzhannikova, A. V., Demonterova, E. I., Jansen, J. D., Preusser, F., & Kamenetsky, M. B. (2018). Catastrophic events in the quaternary outflow history of Lake Baikal. Earth-Science Reviews, 177, 76–113. https://doi.org/10.1016/J.EARSCIREV.2017.11.011.
Avise, J. C., Walker, D., Burns, J. M., Janzen, D. H., & Hallwachs, W. (1999). Species realities and numbers in sexual vertebrates: perspectives from an asexually transmitted genome. Proceedings of the National Academy of Sciences of the United States of America, 96(3), 992–995. https://doi.org/10.1073/pnas.96.3.992.
Berbel-Filho, W. M., Ramos, T. P. A., Jacobina, U. P., Maia, D. J. G., Torres, R. A., & Lima, S. M. Q. (2018). Updated checklist and DNA barcode-based species delimitations reveal taxonomic uncertainties among freshwater fishes from the mid-north-eastern Caatinga ecoregion, north-eastern Brazil. Journal of Fish Biology, 93(2), 311–323. https://doi.org/10.1111/jfb.13758.
Bohlen, J., Šlechtová, V., Bogutskaya, N., & Freyhof, J. (2006). Across Siberia and over Europe: Phylogenetic relationships of the freshwater fish genus Rhodeus in Europe and the phylogenetic position of R. sericeus from the River Amur. Molecular Phylogenetics and Evolution, 40(3), 856–865. https://doi.org/10.1016/j.ympev.2006.04.020.
Bolotov, I. N., Vikhrev, I. V., Bespalaya, Y. V., Gofarov, M. Y., Kondakov, A. V., Konopleva, E. S., Bolotov, N. N., & Lyubas, A. A. (2016). Multi-locus fossil-calibrated phylogeny, biogeography and a subgeneric revision of the Margaritiferidae (Mollusca: Bivalvia: Unionoida). Molecular Phylogenetics and Evolution, 103, 104–121. https://doi.org/10.1016/j.ympev.2016.07.020.
Boo, G. H., & Hughey, J. R. (2019). Phylogenomics and multigene phylogenies decipher two new cryptic marine algae from California, Gelidium gabrielsonii and G. kathyanniae (Gelidiales, Rhodophyta). Journal of Phycology, 55(1), 160–172. https://doi.org/10.1111/jpy.12802.
Corral-Lou, A., Perea, S., Aparicio, E., & Doadrio, I. (2019). Phylogeography and species delineation of the genus Phoxinus Rafinesque, 1820 (Actinopterygii: Leuciscidae) in the Iberian Peninsula. Journal of Zoological Systematics and Evolutionary Research, 57(4), 926–941. https://doi.org/10.1111/jzs.12320.
Crête-Lafrenière, A., Weir, L. K., & Bernatchez, L. (2012). Framing the Salmonidae family phylogenetic portrait: a more complete picture from increased taxon sampling. PLoS ONE, 7(10), e46662. https://doi.org/10.1371/journal.pone.0046662.
Cumbaa, S. L., McAllister, D. E., & Morlan, R. E. (1981). Late Pleistocene fish fossils of Coregonus, Stenodus, Thymallus, Catostomus, Lota, and Cottus from the Old Crow basin, northern Yukon, Canada. Canadian Journal of Earth Sciences, 18(11), 1740–1754. https://doi.org/10.1139/e81-159.
Dawnay, N., Dawnay, L., Hughes, R. N., Cove, R., & Taylor, M. I. (2011). Substantial genetic structure among stocked and native populations of the European grayling (Thymallus thymallus, Salmonidae) in the United Kingdom. Conservation Genetics, 12(3), 731–744. https://doi.org/10.1007/s10592-010-0179-4.
Denys, G. P. J., Secci-Petretto, G., & Gomez dos Santos, A. (2020). The complete mitochondrial genome of Thymallus thymallus (Linnaeus, 1758) (Actinopterygii, Salmonidae) obtained by long range PCRs and double multiplexing. Cybium, 44(2), 91–94. https://doi.org/10.26028/cybium/2020-442-001.
Dierckxsens, N., Mardulyn, P., & Smits, G. (2016). NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Research, 45(4), gkw955. https://doi.org/10.1093/nar/gkw955.
Drummond, A. J., & Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7(1), 214. https://doi.org/10.1186/1471-2148-7-214.
Drummond, A. J., Ho, S. Y. W., Phillips, M. J., & Rambaut, A. (2006). Relaxed phylogenetics and dating with confidence. PLoS Biology, 4(5), e88. https://doi.org/10.1371/journal.pbio.0040088.
Dyldin, Y., Hanel, V., Romanov, V., & Plesnik, J. (2017). A review of the fish genus Thymallus (Pisces: Salmoniformes, Salmonidae, Thymallinae) with taxonomic notes. Bulletin Lampetra, ZO ČSOP Vlašim, 8, 103–126.
ESRI. (2019). ArcGIS desktop: Release 10.7. Redlands, CA: Environmental Systems Research Institute.
Free, C. M., Jensen, O. P., & Mendsaikhan, B. (2015). A mixed-method approach for quantifying illegal fishing and its impact on an endangered fish species. PLoS ONE, 10(12), 1–17. https://doi.org/10.1371/journal.pone.0143960.
Fricke, R., Eschmeyer, W. N. & van der Laan R. (eds.) (2020). Eschmeyer's Catalog of Fishes: Genera, Species, References. (http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp). Electronic version Accessed 15 Feb 2020.
Froufe, E., Alekseyev, S., Knizhin, I., Alexandrino, P., & Weiss, S. (2003a). Comparative phylogeography of salmonid fishes (Salmonidae) reveals late to post-Pleistocene exchange between three now-disjunct river basins in Siberia. Diversity and Distributions, 9(4), 269–282. https://doi.org/10.1046/j.1472-4642.2003.00024.x.
Froufe, E., Knizhin, I., Koskinen, M. T., Primmer, C. R., & Weiss, S. (2003b). Identification of reproductively isolated lineages of Amur grayling (Thymallus grubii Dybowski 1869): concordance between phenotypic and genetic variation. Molecular Ecology, 12, 2345–2355. https://doi.org/10.1046/j.1365-294X.2003.01901.x.
Froufe, E., Knizhin, I., & Weiss, S. (2005). Phylogenetic analysis of the genus Thymallus (grayling) based on mtDNA control region and ATPase 6 genes, with inferences on control region constraints and broad-scale Eurasian phylogeography. Molecular Phylogenetics and Evolution, 34, 106–117. https://doi.org/10.1016/j.ympev.2004.09.009.
Gernhard, T. (2008). The conditioned reconstructed process. Journal of Theoretical Biology, 253(4), 769–778. https://doi.org/10.1016/J.JTBI.2008.04.005.
Gomes-dos-Santos, A., Froufe, E., Gonçalves, D. V., Sousa, R., Prié, V., Ghamizi, M., Benaissa, H., Varandas, S., Teixeira, A., & Lopes-Lima, M. (2019). Freshwater conservation assessments in (semi-) arid regions: Testing river intermittence and buffer strategies using freshwater mussels (Bivalvia, Unionida) in Morocco. Biological Conservation, 236, 420–434. https://doi.org/10.1016/j.biocon.2019.05.038.
Grosswald, M. G. (1998). Late-Weichselian ice sheets in Arctic and Pacific Siberia. Quaternary International, 45–46(97), 3–18. https://doi.org/10.1016/s1040-6182(97)00002-5.
Grosswald, M. G. (2009). Ice sheets in the Russian north and northeast during the last Great Chill. In Data on Glaciological Studies (Vol. 106) Moscow, Nauka, 152 pp. [In Russian].
Gum, B., Gross, R., & Geist, J. (2009). Conservation genetics and management implications for European grayling, Thymallus thymallus: Synthesis of phylogeography and population genetics. Fisheries Management and Ecology, 16(1), 37–51. https://doi.org/10.1111/j.1365-2400.2008.00641.x.
Guo, B., Fang, B., Shikano, T., Momigliano, P., Wang, C., Kravchenko, A., & Merilä, J. (2019). A phylogenomic perspective on diversity, hybridization and evolutionary affinities in the stickleback genus Pungitius. Molecular Ecology, 28(17), 4046–4064. https://doi.org/10.1111/mec.15204.
Hahn, C., Bachmann, L., & Chevreux, B. (2013). Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Research, 41(13), e129–e129. https://doi.org/10.1093/nar/gkt371.
He, B., Su, T., Wu, Y., Xu, J., & Huang, D. (2018). Phylogenetic analysis of the mitochondrial genomes in bees (Hymenoptera: Apoidea: Anthophila). PLoS ONE, 13(8), e0202187. https://doi.org/10.1371/journal.pone.0202187.
Hebert, P. D. N., Penton, E. H., Burns, J. M., Janzen, D. H., & Hallwachs, W. (2004). Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences of the United States of America, 101(41), 14812–14817. https://doi.org/10.1073/pnas.0406166101.
Huml, V. J., Taylor, M. I., Edwin Harris, W., Sen, R., & Ellis, J. S. (2018). Neutral variation does not predict immunogenetic variation in the European grayling (Thymallus thymallus) - implications for management. Molecular Ecology, 27(21), 4157–4173. https://doi.org/10.1111/mec.14864.
Iwasaki, W., Fukunaga, T., Isagozawa, R., Yamada, K., Maeda, Y., Satoh, T., et al. (2013). MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Molecular Biology and Evolution, 30(11), 2531–2540. https://doi.org/10.1093/molbev/mst141.
Kapli, P., Lutteropp, S., Zhang, J., Kobert, K., Pavlidis, P., Stamatakis, A., & Flouri, T. (2017). Multi-rate Poisson Tree Processes for single-locus species delimitation under maximum likelihood and Markov Chain Monte Carlo. Bioinformatics, 33, 1630–1638. https://doi.org/10.1093/bioinformatics/btx025.
Katoh, K., & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772–780. https://doi.org/10.1093/molbev/mst010.
Kaus, A., Michalski, S., Hänfling, B., Karthe, D., Borchardt, D., & Durka, W. (2019). Fish conservation in the land of steppe and sky: evolutionarily significant units of threatened salmonid species in Mongolia mirror major river basins. Ecology and Evolution, 9(6), 3416–3433. https://doi.org/10.1002/ece3.4974.
Kirillov, A. F., & Knizhin, I. B. (2014). Ichthyofauna of the Lena River (Laptev Sea Basin): modern composition and historical formation. Journal of Ichthyology, 54(7), 433–445. https://doi.org/10.1134/S0032945214040031.
Knizhin, I. B., & Weiss, S. J. (2009). A new species of grayling Thymallus svetovidovi sp. nova (Thymallidae) from the Yenisei basin and its position in the genus Thymallus. Journal of Ichthyology, 49(1), 1–9. https://doi.org/10.1134/S0032945209010019.
Knizhin, I. B., Weiss, S. J., Antonov, A. L., & Froufe, E. (2004). Morphological and genetic diversity of Amur graylings (Thymallus, Thymallidae). Journal of Ichthyology, 44(1), 52–69.
Knizhin, I. B., Antonov, A. L., & Weiss, S. J. (2006a). A new subspecies of the Amur grayling Thymallus grubii flavomaculatus ssp. nova (Thymallidae). Journal of Ichthyology, 46(8), 555–562. https://doi.org/10.1134/S0032945206080017.
Knizhin, I. B., Weiss, S. J., & Sušnik, S. (2006b). Graylings of Baikal Lake basin (Thymallus, Thymallidae): diversity of forms and their taxonomic status. Journal of Ichthyology, 46(6), 418–435. https://doi.org/10.1134/S0032945206060026.
Knizhin, I. B., Weiss, S. J., Bogdanov, B. E., Samarina, S. S., & Froufe, E. (2006c). Finding a new form of the grayling Thymallus arcticus (Thymallidae) in the basin of Lake Baikal. Journal of Ichthyology, 46, 34–43. https://doi.org/10.1134/S003294520601005X.
Knizhin, I. B., Antonov, A. L., Safronov, S. N., & Weiss, S. J. (2007). New species of grayling Thymallus tugarinae sp. nova (Thymallidae) from the Amur River basin. Journal of Ichthyology, 47(2), 123–139. https://doi.org/10.1134/S0032945207020014.
Knizhin, I. B., Weiss, S. J., Bogdanov, B. E., Kopun, T., & Muzalevskaya, O. V. (2008). Graylings (Thymallidae) of water bodies in western Mongolia: morphological and genetic diversity. Journal of Ichthyology, 48(9), 714–735. https://doi.org/10.1134/S0032945208090038.
Komatsu, G., Baker, V. R., Arzhannikov, S. G., Gallagher, R., Arzhannikova, A., & Oguchi, T. (2016). Catastrophic flooding, palaeolakes, and late Quaternary drainage reorganization in northern Eurasia. International Geology Review, 58(14), 1693–1722. https://doi.org/10.1080/00206814.2015.1048314.
Koskinen, M. T., Knizhin, I., Primmer, C. R., Schlötterer, C., & Weiss, S. (2002). Mitochondrial and nuclear DNA phylogeography of Thymallus spp. (grayling) provides evidence of ice-age mediated environmental perturbations in the world’s oldest body of fresh water, Lake Baikal. Molecular Ecology, 11(12), 2599–2611. https://doi.org/10.1046/j.1365-294X.2002.01642.x.
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547–1549. https://doi.org/10.1093/molbev/msy096.
Lanfear, R., Calcott, B., Ho, S. Y. W., & Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution, 29(6), 1695–1701. https://doi.org/10.1093/molbev/mss020.
Lazarev, S., de Leeuw, A., Stoica, M., Mandic, O., van Baak, C. G. C., Vasiliev, I., & Krijgsman, W. (2020). From Khersonian drying to Pontian “flooding”: late Miocene stratigraphy and palaeoenvironmental evolution of the Dacian Basin (Eastern Paratethys). Global and Planetary Change, 192, 103224. https://doi.org/10.1016/j.gloplacha.2020.103224.
Lecaudey, L. A., Schliewen, U. K., Osinov, A. G., Taylor, E. B., Bernatchez, L., & Weiss, S. J. (2018). Molecular phylogenetics and evolution inferring phylogenetic structure, hybridization and divergence times within Salmoninae (Teleostei : Salmonidae) using RAD-sequencing. Molecular Phylogenetics and Evolution, 124, 82–99. https://doi.org/10.1016/j.ympev.2018.02.022.
Lemey, P., Rambaut, A., Drummond, A. J., & Suchard, M. A. (2009). Bayesian phylogeography finds its roots. PLoS Computational Biology, 5(9). https://doi.org/10.1371/journal.pcbi.1000520.
Liedigk, R., Kolleck, J., Böker, K. O., Meijaard, E., Md-Zain, B. M., Abu, M., et al. (2015). Mitogenomic phylogeny of the common long-tailed macaque (Macaca fascicularis fascicularis). BMC Genomics, 16(1), 222. https://doi.org/10.1186/s12864-015-1437-0.
Linke, S., Lehner, B., Ouellet Dallaire, C., Ariwi, J., Grill, G., Anand, M., et al. (2019). Global hydro-environmental sub-basin and river reach characteristics at high spatial resolution. Scientific Data, 6(1), 283. https://doi.org/10.1038/s41597-019-0300-6.
Lopes-Lima, M., Burlakova, L., Karatayev, A., Gomes-dos-Santos, A., Zieritz, A., Froufe, E., & Bogan, A. E. (2019). Revisiting the North American freshwater mussel genus Quadrula sensu lato (Bivalvia Unionidae): phylogeny, taxonomy and species delineation. Zoologica Scripta, 48(3), 313–336. https://doi.org/10.1111/zsc.12344.
Ma, B., Jiang, H., Sun, P., Chen, J., Li, L., Zhang, X., & Yuan, L. (2016). Phylogeny and dating of divergences within the genus Thymallus (Salmonidae: Thymallinae) using complete mitochondrial genomes. Mitochondrial DNA Part A, 27(5), 3602–3611. https://doi.org/10.3109/19401736.2015.1079824.
Mäkinen, H., Papakostas, S., Vøllestad, L. A., Leder, E. H., & Primmer, C.R. (2016). Plastic and evolutionary gene expression responses are correlated in European grayling (Thymallus thymallus) subpopulations adapted to different thermal environments. Journal of Heredity, 107, 82–89. https://doi.org/10.1093/jhered/esv069.
Marić, S., Kalamujić, B., Snoj, A., Razpet, A., Lukić-Bilela, L., Pojskić, N., & Sušnik Bajec, S. (2012). Genetic variation of European grayling (Thymallus thymallus) populations in the Western Balkans. Hydrobiologia, 691(1), 225–237. https://doi.org/10.1007/s10750-012-1076-2.
Marić, S., I. V., Askeyev, O. V., Askeyev, S. P., Monakhov, J. B., & Snoj, A. (2014). Phylogenetic and population genetic analysis of Thymallus thymallus (Actinopterygii, Salmonidae) from the middle Volga and upper Ural drainages. Hydrobiologia, 740, 167–176.
McAllister, D. E., & Harington, C. R. (1969). Pleistocene grayling, Thymallus , from Yukon, Canada. Canadian Journal of Earth Sciences, 6(5), 1185–1190. https://doi.org/10.1139/e69-119.
Meraner, A., Cornetti, L., & Gandolfi, A. (2014). Defining conservation units in a stocking-induced genetic melting pot: unraveling native and multiple exotic genetic imprints of recent and historical secondary contact in Adriatic grayling. Ecology and Evolution, 4(8), 1313–1327. https://doi.org/10.1002/ece3.931.
Miller, M. A., Pfeiffer, W., & Schwartz, T. (2010). Creating the CIPRES science gateway for inference of large phylogenetic trees. In 2010 Gateway Computing Environments Workshop (GCE) (pp. 1–8). IEEE. https://doi.org/10.1109/GCE.2010.5676129.
Miller, J. M., Malenfant, R. M., Moore, S. S., & Coltman, D. W. (2012). Short reads, circular genome: skimming SOLiD sequence to construct the Bighorn sheep mitochondrial genome. Journal of Heredity, 103(1), 140–146. https://doi.org/10.1093/jhered/esr104.
Miya, M., & Nishida, M. (2015). The mitogenomic contributions to molecular phylogenetics and evolution of fishes: a 15-year retrospect. Ichthyological Research, 62(1), 29–71. https://doi.org/10.1007/s10228-014-0440-9.
Mueller, M., Pander, J., & Geist, J. (2018). Comprehensive analysis of >30 years of data on stream fish population trends and conservation status in Bavaria, Germany. Biological Conservation, 226, 311–320. https://doi.org/10.1016/j.biocon.2018.08.006.
Ocock, J., Baasanjav, G., Baillie, J. E. M., Erbenebat, M., Kottelat, M., Mendsaikhan, B., & Smith, K. (2006). Mongolian red list of fishes. London, UK: Zoological Society of London.
Olson, K. W., Krabbenhoft, T. J., Hrabik, T. R., Mendsaikhan, B., & Jensen, O. P. (2019). Pelagic–littoral resource polymorphism in Hovsgol grayling Thymallus nigrescens from Lake Hovsgol, Mongolia. Ecology of Freshwater Fish, 28(3), 411–423. https://doi.org/10.1111/eff.12464.
Osinov, A. G., & Lebedev, V. S. (2000). Genetc divergence and phylogeny of the Salmoninae based on allozyme data. Journal of Fish Biology, 57(2), 354–381. https://doi.org/10.1111/j.1095-8649.2000.tb02177.x.
Palcu, D. V., Vasiliev, I., Stoica, M., & Krijgsman, W. (2019). The end of the Great Khersonian Drying of Eurasia: magnetostratigraphic dating of the Maeotian transgression in the Eastern Paratethys. Basin Research, 31(1), 33–58. https://doi.org/10.1111/bre.12307.
Pan, T., Sun, Z., Lai, X., Orozcoterwengel, P., Yan, P., & Wu, G…Zhang, B. (2019). Hidden species diversity in Pachyhynobius: a multiple approaches species delimitation with mitogenomes. Molecular Phylogenetics and Evolution, 137, 138–145. https://doi.org/10.1016/J.YMPEV.2019.05.005.
Papakostas, S., Vøllestad, L. A., Bruneaux, M., Aykanat, T., Vanoverbeke, J., Ning, M., et al. (2014). Gene pleiotropy constrains gene expression changes in fish adapted to different thermal conditions. Nature Communications, 5(1), 4071. https://doi.org/10.1038/ncomms5071.
Patil, T. S., Jamdade, R. A., Patil, S. M., Govindwar, S. P., & Muley, D. V. (2018). DNA barcode based delineation of freshwater fishes from northern Western Ghats of India, one of the world’s biodiversity hotspots. Biodiversity and Conservation, 27(13), 3349–3371. https://doi.org/10.1007/s10531-018-1604-0.
Perdices, A., Bohlen, J., Šlechtová, V., & Doadrio, I. (2016). Molecular evidence for multiple origins of the European spined loaches (Teleostei, Cobitidae). PLoS ONE, 11(1), 1–16. https://doi.org/10.1371/journal.pone.0144628.
Persat, H., Mattersdorfer, K., Charlat, S., Schenekar, T., & Weiss, S. (2016). Genetic integrity of the European grayling (Thymallus thymallus) populations within the Vienne River drainage basin after five decades of stockings. Cybium, 40(1), 7–20.
Persat, H., Weiss, S., Froufe, E., Secci-Petretto, G., & Denys, G. P. (2019). A third European species of grayling (Actinopterygii, Salmonidae), endemic to the Loire River basin (France), Thymallus ligericus n. sp. Cybium, 43(3), 233–238.
Puillandre, N., Lambert, A., Brouillet, S., & Achaz, G. (2012). ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology, 21(8), 1864–1877. https://doi.org/10.1111/j.1365-294X.2011.05239.x.
de Queiroz, K. (2007). Species concepts and species delimitation. Systematic Biology, 56(6), 879–886. https://doi.org/10.1080/10635150701701083.
Rambaut A. (2012). FigTree v1. 4. Molecular evolution, phylogenetics and epidemiology. Edinburgh, UK: Retrieved from http://tree.bio.ed.ac.uk/software/figtree.
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., & Suchard, M. A. (2018). Posterior summarization in Bayesian phylogenetics using tracer 1.7. Systematic Biology, 67(5), 901–904. https://doi.org/10.1093/sysbio/syy032.
Ratnasingham, S., & Hebert, P. D. (2007). BARCODING: bold: the barcode of life data system (http://www.barcodinglife.org). Molecular Ecology Notes, 7(3), 355–364. https://doi.org/10.1111/j.1471-8286.2007.01678.x.
Ritchie, A. M., Lo, N., & Ho, S. Y. W. (2016). Examining the sensitivity of molecular species delimitations to the choice of mitochondrial marker. Organisms, Diversity and Evolution, 16(3), 467–480. https://doi.org/10.1007/s13127-016-0275-5.
Roman, I., Bourgeois, Y., Reyes-Velasco, J., Jensen, O. P., Waldman, J., & Boissinot, S. (2018). Contrasted patterns of divergence and gene flow among five fish species in a Mongolian rift lake following glaciation. Biological Journal of the Linnean Society, 125, 115–125. https://doi.org/10.1093/biolinnean/bly083/5051965.
Ronquist, F., & Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19(12), 1572–1574. https://doi.org/10.1093/bioinformatics/btg180.
Rückert-Ülkümen, N., & Kaya, O. (1993). Neue Beiträge zur Tertiär-Stratigraphie und Otolithenfauna der Umgebung von Istanbul (Küçükçekmece-Büyükçekmece See), Türkei. Mitteilungen. Bayerische Staatssammlung für Paläontologie und Historische Geologie, 33, 51–89 [in German].
Rylková, K., Kalous, L., Bohlen, J., Lamatsch, D. K., & Petrtýl, M. (2013). Phylogeny and biogeographic history of the cyprinid fish genus Carassius (Teleostei: Cyprinidae) with focus on natural and anthropogenic arrivals in Europe. Aquaculture, 380–383, 13–20. https://doi.org/10.1016/j.aquaculture.2012.11.027.
Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Extraction and purification of plasmid DNA. Molecular Cloning: A Laboratory Manual, 1(1), 21–152.
Sävilammi, T., Primmer, C. R., Varadharajan, S., Guyomard, R., Guiguen, Y., Sandve, S. R., Vøllestad, L. A., Papakostas, S., & Lien, S. (2019). The chromosom-level genome assembly of European grayling reveals aspects of a unique genome evolution process within salmonds. G3: Genes, Genomes, Genetics, 9(5), 1283–1294. https://doi.org/10.1534/g3.118.200919.
Skog, A., Vøllestad, L. A., Stenseth, N. C., Kasumyan, A., & Jakobsen, K. S. (2014). Circumpolar phylogeography of the northern pike (Esox lucius) and its relationship to the Amur pike (E. reichertii). Frontiers in Zoology, 11(1), 1–8. https://doi.org/10.1186/s12983-014-0067-8.
Skurikhina, L. A., Mednikov, B. M., & Tugarina, P. Y. (1985). Geneticheskaya divergentziya khariusov (Thymallus Cuvier) Evrazii i seti vidov. Zoologicheskiĭ Zhurnal, 64(2), 245 [In Russian].
Šlechtová, V., Bohlen, J., & Perdices, A. (2008). Molecular phylogeny of the freshwater fish family Cobitidae (Cypriniformes: Teleostei): delimitation of genera, mitochondrial introgression and evolution of sexual dimorphism. Molecular Phylogenetics and Evolution, 47(2), 812–831. https://doi.org/10.1016/j.ympev.2007.12.018.
Smith, G. R. (1992). Introgression in fishes: significance for paleontology, cladistics, and evolutionary rates. Systematic Biology, 41(1), 41–57. https://doi.org/10.1093/sysbio/41.1.41.
Srivathsan, A., & Meier, R. (2012). On the inappropriate use of Kimura-2-parameter (K2P) divergences in the DNA-barcoding literature. Cladistics, 28(2), 190–194. https://doi.org/10.1111/j.1096-0031.2011.00370.x.
Stamatakis, A. (2016). The RAxML v8.2.X manual. Retrieved from http://sco.h-its.org/exelixis/web/software/raxml/index.html
Stamford, M. D., & Taylor, E. B. (2004). Phylogeographical lineages of Arctic grayling (Thymallus arcticus) in North America: divergence, origins and affinities with Eurasian Thymallus. Molecular Ecology, 13(6), 1533–1549. https://doi.org/10.1111/j.1365-294X.2004.02174.x.
Suchard, M. A., & Rambaut, A. (2009). Many-core algorithms for statistical phylogenetics. Bioinformatics, 25(11), 1370–1376. https://doi.org/10.1093/bioinformatics/btp244.
Sukumaran, J., & Knowles, L. L. (2017). Multispecies coalescent delimits structure, not species. Proceedings of the National Academy of Sciences of the United States of America, 114(7), 1607–1612. https://doi.org/10.1073/pnas.1607921114.
Sušnik, S., Berrebi, P., Dovč, P., Hansen, M. M., & Snoj, A. (2004). Genetic introgression between wild and stocked salmonids and the prospects for using molecular markers in population rehabilitation: The case of the Adriatic grayling (Thymallus thymallus L. 1785). Heredity, 93(3), 273–282. https://doi.org/10.1038/sj.hdy.6800500.
Van Houdt, J. K. J., De Cleyn, L., Perretti, A., & Volckaert, F. A. M. (2005). A mitogenic view on the evolutionary history of the Holarctic freshwater gadoid, burbot (Lota lota). Molecular Ecology, 14(8), 2445–2457. https://doi.org/10.1111/j.1365-294X.2005.02590.x.
Varadharajan, S., Sandve, S. R., Gillard, G. B., Tørresen, O. K., Mulugeta, T. D., Hvidsten, T. R., et al. (2018). The grayling genome reveals selection on gene expression regulation after whole genome duplication. Genome Biology and Evolution, 10(10), 2785–2800. https://doi.org/10.1093/gbe/evy201.
Ward, R. D. (2009). DNA barcode divergence among species and genera of birds and fishes. Molecular Ecology Resources, 9(4), 1077–1085. https://doi.org/10.1111/j.1755-0998.2009.02541.x.
Weiss, S., Persat, H., Eppe, R., Schlötterer, C., & Uiblein, F. (2002). Complex patterns of colonization and refugia revealed for European grayling Thymallus thymallus, based on complete sequencing of the mitochondrial DNA control region. Molecular Ecology, 11(8), 1393–1407. https://doi.org/10.1046/j.1365-294X.2002.01544.x.
Weiss, S., Knizhin, I., Kirillov, A., & Froufe, E. (2006). Phenotypic and genetic differentiation of two major phylogeographical lineages of arctic grayling Thymallus arcticus in the Lena River, and surrounding Arctic drainages. Biological Journal of the Linnean Society, 88(4), 511–525. https://doi.org/10.1111/j.1095-8312.2006.00621.x.
Weiss, S., Knizhin, I., Romanov, V., & Kopun, T. (2007). Secondary contact between two divergent lineages of grayling Thymallus in the lower Enisey basin and its taxonomic implications. Journal of Fish Biology, 71(SUPPL. C), 371–386. https://doi.org/10.1111/j.1095-8649.2007.01662.x.
Weiss, S. J., Kopun, T., & Sušnik Bajec, S. (2013). Assessing natural and disturbed population structure in European grayling Thymallus thymallus: Melding phylogeographic, population genetic and jurisdictional perspectives for conservation planning. Journal of Fish Biology, 82(2), 505–521. https://doi.org/10.1111/jfb.12007.
Weiss, S., Grimm, J., Gonçalves, D. V., Secci-Petretto, G., Englmaier, G. K., Baimukanov, M., & Froufe, E. (2020a). Comparative genetic analysis of grayling (Thymallus spp. Salmonidae) across the paleohydrologically dynamic river drainages of the Altai-Sayan mountain region. Hydrobiologia, 847, 2823–2844. https://doi.org/10.1007/s10750-020-04273-3.
Weiss, S., Secci-Petretto, G., Antonov, A., & Froufe, E. (2020b). Multiple species of grayling (Thymallus sp.) found in sympatry in a remote tributary of the Amur River. Zoologica Scripta, 49(1), 117–128. https://doi.org/10.1111/zsc.12393.
Zhang, J., Kapli, P., Pavlidis, P., & Stamatakis, A. (2013). A general species delimitation method with applications to phylogenetic placements. Bioinformatics, 29(22), 2869–2876. https://doi.org/10.1093/bioinformatics/btt499.
Funding
Open access funding provided by University of Graz. This work was supported by the Foundation for Science and Technology to GS (SFRH/BD/139069/2018) and to AGS (SFRH/BD/137935/2018), by Strategic Funding UIDB/04423/2020 and UIDP/04423/2020 and by the Institute of Biology at the University of Graz in the form of a Doc Academy Grant to GE under the supervision of SW. DVG is supported by project ConBiOmics (NORTE-01-0145-FEDER-030286, contract IA/2018–079). The 14 mitogenomes sequenced in France were done according to the partnership MNHN-AFB 2016–2018 and with the assistance of the ‘Service de Systématique Moléculaire’ of the Muséum national d’Histoire naturelle (CNRS UMS 2700).
Author information
Authors and Affiliations
Contributions
SW and EF conceived the study for this manuscript with input from DVG. GS, AGS, GD, CH, AA, HP and GE collected data and resources, and helped construct range distribution maps. Most raw data analysis was performed by GS, DVG, AGS and EF. SW wrote the first draft, and all authors contributed to writing.
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
Mitogenome map of Thymallus. Gene order was identical in all individuals. The genes represented outside the circle are encoded in the light strand; the genes represented inside the circle are encoded on the heavy strand. Red: tRNA; Black: protein-coding genes; Light brown: rRNA; Dark brown: control region. (PNG 285 kb)
ESM 2
Frequency histograms of the p distances calculated between the putative species and between the MOTUs defined in each of the three species delineation methods applied. Gaps between frequency peaks illustrate gaps between levels of phylogenetic structure. (JPG 62 kb)
ESM 3
Box plot of pairwise divergences for the whole mitogenome (excluding the CR) and all protein-coding genes: a) based on uncorrected p distances, and b) based on K2P distances. (PDF 1430 kb)
ESM 4
List of mitogenomes of all species used in the analyses. Shown are the species name and common name, GenBank accession numbers, the sample location and major basin, the country and the corresponding reference. (XLSX 22 kb)
ESM 5
List of samples processed with Illumina low-coverage genome-wide sequencing. Shown are the BioSample, and SRA accession numbers, the total number of genomic reads, the total number of mitochondrial reads, the percent of total reads that were mitochondrial, and the average organelle coverage across the mitogenome. (DOCX 15 kb)
ESM 6
Pairwise uncorrected net p-distances of the whole mitogenome (excl. CR) among species. (DOCX 18 kb)
ESM 7
Expanded taxonomic review of all taxa evaluated in this study as well as notes on some potential synonymies or unevaluated taxa, historical notes and justification for supporting status as a species as well as suggested global conservation status. (DOCX 59.3 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Weiss, S.J., Gonçalves, D.V., Secci-Petretto, G. et al. Global systematic diversity, range distributions, conservation and taxonomic assessments of graylings (Teleostei: Salmonidae; Thymallus spp.). Org Divers Evol 21, 25–42 (2021). https://doi.org/10.1007/s13127-020-00468-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-020-00468-7