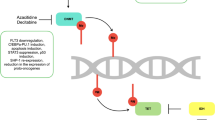
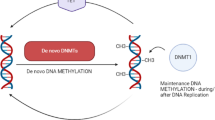
Abstract
Background
Fragile histidine triad (FHIT) has been documented to play a vital role in various cancers including acute lymphoblastic leukemia (ALL). Keeping in view the plausible role of FHIT gene, we aimed to examine DNA promoter hypermethylation and mRNA expression in ALL cases in Kashmir (North India).
Methods
A total of 66 cases of ALL were analyzed for FHIT mRNA expression and promoter methylation by qRT-PCR and Methylation Specific-PCR (MS-PCR) respectively.
Results
FHIT mRNA expression showed significantly decreased expression in ALL cases with mean fold change of 9.24 ± 5.44 as compared to healthy controls (p = 0.01). The pattern of FHIT deregulation in ALL cases differed significantly between decreased and increased expression (p < 0.0001). A threefold decreased expression was observed in 75% of ALL cases than healthy controls (− 3.58 ± 2.32). ALL patients with FHIT gene promoter hypermethylation presented significantly higher in 80% (53/66) of cases (p = 0.0005). The association of FHIT gene hypermethylation and its subsequent expression showed FHIT mRNA expression as significantly lower in ALL cases with hypermethylation (p = 0.0008). B-ALL cases exhibited a highly significant association between the methylation pattern and its mRNA expression (p = 0.000). In low range WBC group, a significant association was found between increased expression (26%) of the cases and methylated (4%)/unmethylated group 86% (p = 0.0006).
Conclusion
The present study conclude that FHIT gene hypermethylation and its altered expression may be linked in the pathogenesis of ALL and provide an evidence for the role of FHIT in the development of ALL.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Among different types of leukemia, acute lymphoblastic leukemia (ALL) is the most common form of childhood cancer. As per Surveillance, Epidemiology, and End Results (SEER) program, ALL accounts for ~ 25% of all diagnosed cancers in children up to the age group of 14 years [1]. Here in this region (J&K, India), ALL ranks 5th among most common cancers and occurs at a frequency of 9.9% and the average incidence of leukemia is 5.8/105/year with highest incidence in ALL cases [2]. As per the latest report, Leukemia is the most prevalent childhood cancer that accounts for 36.6% of cases and ALL was the most common type found than any other subtypes [3]. While 80% of ALL occurs in children, it is a more devastating disease in adults. Among ALL, Precursor B-cell ALL (BCP-ALL) represents 73–85% of total cases. B-ALL is generally related with a better outcome in children where the rate of cure touches ~ 90% [4]. Despite refinement for improved treatment response, ALL still poses a challenge as a leading cause of cancer-related mortality due to relapse [5]. Several studies have investigated the progression of genetic alterations from diagnosis to relapse that contribute to drug resistance and the assumption is that identifying the basic biological mechanisms that are accountable for drug resistance/relapse would offer rational resources for both inhibition of relapse and successful salvage therapy [6, 7]. Analyses of DNA copy number mutations, gene expression, DNA methylation, and next-generation sequencing of ALL samples in addition to chromosomal aberrations have been used to classify global genetic and epigenetic changes that characterize disease progression [8,9,10,11]. There are specific established risk factors in ALL that primarily include gender, age, performance score, leukocyte count, cytogenetics and molecular alterations and the immunophenotyping etc. ([12]). Chromosomal aberrations are common characteristics in ALL and the fragile histidine triad (FHIT) gene is a main spot of chromosomal rearrangement [13]. The FHIT gene acts as tumor suppressive gene and its tumor suppressive properties have been shown to be restored by transfection of FHIT in FHIT‑deficient human cancer cells that emerge to stimulate apoptosis and hamper cellular growth [14, 15]. FHIT promoter hypermethylation and its expression has been observed as one of the key events in the pathogenesis of ALL. Promoter DNA methylation, an epigenetic modification is notably seen to contribute to the development of leukemia as confirmed in case of FHIT gene in various forms of cancers [16,17,18] and in particular ALL [19, 20]. Pathogenesis of many leukemic disorders has been associated with FHIT loss of function as a consequence of its down regulation or aberrant expression [21]. In ALL, loss of FHIT expression causes inactivation of FHIT protein that may result in the development of leukemias [22]. Keeping in view the plausible role of FHIT gene, we conducted this study to observe the pattern and significance of FHIT gene promoter methylation and its expression in the pathogenesis of ALL with respect to clinic-pathological characteristics.
2 Methods
The present study was conducted in Advanced Center for Human Genetics, Sher-i-Kashmir institute of Medical Sciences (SKIMS), Srinagar (J&K, India). A total of 66 ALL patients were recruited between 2018 and 2021 who attended the Department of Clinical Hematology and Medical Oncology (SKIMS).. The diagnosis of ALL was confirmed by clinical assessment along with bone marrow and cytochemical cytopathology. ALL patients were categorized according to the French & British American classification (FAB) and WHO classification of acute leukemia. ALL patients were classified into different set of risk groups and treatment was given as per the modified BFM-95 protocol [23]. Peripheral blood samples (2–3 ml) were taken from all the patients and from age and sex matched healthy controls with no signs of any hematological/and or any other malignancy from the Out Patients Department of SKIMS. The samples were immediately extracted for DNA and RNA analysis and stored at – 80 ℃ for further processing. The local ethics committee of SKIMS approved the study and written informed consent was obtained from cases (parent consent in case of minor) as well as controls. All patients and guardians were told about the research protocol and blood samples were taken only after signing a previous approval by their patients or guardians. Data of ALL patients (medical records and hematology/bone marrow reports) was obtained from personal interviews with patients and or guardians.
2.1 DNA extraction
Blood samples from ALL patients were used for DNA extraction through phenol/chloroform method and/or DNA extraction kit (Qaigen, Germany). DNA concentration was estimated for purity at the absorbance of 260/280 nm and verified on 0.8–1% agarose gel.
2.2 Analysis of FHIT gene hypermethylation by MS-PCR
Around 1.5–2 µg of DNA extracted was modified with bisulfite modification treatment (EZ-DNA Methylation Kit, Zymo Research Corporation, USA). A methylation specific-PCR (MSP-PCR) was done as per the protocol published by Golam Reza et al. [24]. The gel pictures were analyzed for bands by gel documentation system Flourchem HD2 (Cell Bioscience, USA).
2.3 Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) for FHIT mRNA analysis
From fresh blood samples using TRIZOL reagent (Ambion, Thermo Fisher, USA), RNA was extracted as per the manufacturer’s protocol. The concentration of RNA was examined using a UV–Vis Spectrophotometer (Thermo Scientific,Nano Drop 2000) with a wavelength of 260 nm. Integrity of RNA was checked using 1% Agarose gel (Additional file 1: Figure S1). The DNase, RNase- free (Thermo Scientific, USA) reagent kit was used to remove the traces of genomic DNA from RNA preparation and the Thermo Scientific™ RevertAid™ First Strand cDNA Synthesis Kit (Thermo Scientific, USA) was used for cDNA synthesis as per the protocol using primers as shown in Additional file Table S1. For qRT-PCR of FHIT mRNA expression, the PCR amplification was performed on Rotor-Gene Q accessories (Qiagen, Germany) using Maxima SyBR Green /ROX qPCR Master Mix (2x) with an internal control gene GAPDH as per the protocol (primers: Additional file Table S1). The cycle threshold (Ct) was analyzed and fold changes of gene of interest relative to GAPDH were done by the formula of 2−ΔΔCt. The results were finally presented as an average log2 fold change which represented a decrease or increase in the mRNA expression of the respective gene.
2.4 Statistical analysis
Statistical analysis for the results of the study was done using IBM Statistics SPSS software, version-23 (IBM Corp, NY, USA). For clinico-pathological characteristics, categorical variables (gender and age) in case and controls were matched using chi-squared test. Expression analysis FHIT gene was expressed as cycle threshold values. Comparative analysis was performed Livak Method (ΔΔCt) where data interpretation was exhibited in terms of relative fold expression.
3 Results
FHIT gene methylation was conducted by MS-PCR (n = 66) while as analysis of FHIT mRNA expression levels were conducted in corresponding 40 cases of ALL (25 males and 15 females; mean age, of 12.2 ± 3.4 years; with an age range, of 1–27 years) and 40 healthy (age and sex matched) children 30 males and 12 females; mean age, of 12.3 ± 4.1 years; with an age range, of 1–30 years). The clinic-pathological characteristics of the studied subjects are given in Table 1.
ALL patient samples analyzed for FHIT gene promoter hypermethylation showed significantly higher frequency of hypermethylation in 80% of cases (53/66) (p = 0.0005; Table 1).
FHIT mRNA expression showed significantly decreased expression in 75% of ALL cases with mean fold change of 9.24 ± 5.44 as compared to healthy controls (p = 0.01: Fig. 1). The pattern of altered lower expression was significantly more in ALL cases than controls (p < 0.0001: Table 2). Relative expression of FHIT gene was successfully analyzed in all samples and melt curve analysis was also performed with every experiment which revealed there is stability with no non-specificity within the reaction (Additional file Fig. 2A and B). Lower expression was observed in 75% (30/40) of ALL cases as compared to healthy controls with mean log2 fold change of − 3.58 ± 2.32 that signifies threefold decreased expression (Table 2). Overall 25% (n = 10) cases showed increased expression with an average log2 fold change 2.74 ± 1.40 which signifies 2-fold increased expression of FHIT gene in the cases than controls. However, high significance was observed when decreased expression of ALL cases was compared with the cases having increased expression (Fig. 2B). Among the clinico-pathological characteristics of the patients, FHIT gene expression depicted significant difference in gender (p < 0.013) and rest of the parameters showed no association (p > 0.05) (Table 1).
3.1 Correlation of FHIT gene promoter hypermethylation with its expression
The association of FHIT gene promoter hypermethylation and its subsequent expression were performed on previously obtained samples. The relative expression analysis of all the cases of ALL is shown in Fig. 2A. Of the 30 samples with decreased FHIT gene expression, 90% (27/30) were hypermethylated, while increased expression exhibited 20% (02/10) as methylated sequences (Table 3 and Fig. 2B). The results showed that FHIT mRNA expression was significantly lower in ALL cases with hypermethylation (p = 0.0008). The association of FHIT hypermethylation and its expression in different clinical parameters of ALL patients are shown in (Table 3).
Among immunophenotypes of ALL, B-ALL showed concordant pattern between hypermethylation and mRNA expression. Decreased FHIT expression was observed in 74.2% (26/35) wherein methylated DNA sequences accounted for 68.5% (24/35). Cases with increased mRNA expression of FHIT gene was observed in 25.7% (9/35) wherein 2.8% (1/35) showed hypermethylation. In comparison to T-ALL, a highly significant association was observed between the pattern of methylation and its mRNA expression with B-ALL cases (p = 0.000: Fig. 3A).
Among pediatric age group (< 18 years), 86% (18/21) showed decreased FHIT mRNA expression and accounted for 81% (21/26) of the methylated cases. Increased FHIT expression was detected in 19% (05/26) of the cases with 20% (01/5) methylated cases (Additional file Figure S3).
In low range WBC (< 10 × 109/L) group, FHIT expression was decreased in 74% (20/27) of the cases where 90% (18/20) showed hypermethylated sequences. When compared to rest of the cases in low range WBC group, a significant association was observed between increased expression (26%) of the cases and methylated (4%)/unmethylated group (86%) (p = 0.0006: Table 3 and Fig. 3B). FHIT mRNA expression level for both lower platelet and higher group of ALL patients were significantly decreased in methylated cases, as compared to increased expression observed in unmethylated cases. In lower platelet groups, highly significant association was observed between the methylation status and its mRNA expression among the ALL cases (p = 0.0002). Fusion transcripts were detected in 32.5% (13/40) of the cases and the frequency of these transcripts were as BCR-ABL (n = 4), ETV-RUNX1 (n = 6), MLL-AF4 (n = 2) and TCF-PBX1 (n = 1). When positive fusion transcripts were correlated with FHIT gene hypermethylation and its subsequent expression, 85% (11/13) cases showed decreased expression amongst which 77%(10/13) belong to methylated cases. However, association between the status of fusion transcript cases to expression and methylation did not reveal any significance (Table 3). A Comparison of fold change expression of FHIT gene with clinical parameters is depicted in Additional file 1: Figure S3
4 Discussion
FHIT gene has been associated with a wide range of tumor-suppressive properties like down-regulation of oncogene activity, apoptosis, invasion, and metastasis [25]. Thus any change in FHIT protein expression might result in a number of changes at molecular level and hence it’s altered functions ultimately lead to different kinds of diseases including tumorogenesis [26]. Reports are also available regarding the FHIT gene hypermethylation as an important contributor in causation of different malignancies [27]. Because epigenetic modifications can have significant effect on transcriptional activity in candidate tumor suppressor genes, it can promote tumorogenesis including leukemia’s [28, 29].
4.1 FHIT gene mRNA expression and its association with FHIT promoter hyper-methylation
Recent advances in the molecular studies have led to deep understanding of ALL and a number of studies have provided evidence for the constitutive activation of FHIT gene and its association with ALL [16, 17]. Altered expression of FHIT is one of the potential mechanisms that promote tumor progression and its decreased expression is associated with signaling pathways in various cancers [30].
In the present study, the prognostic significance of FHIT expression in ALL patients was evaluated for FHIT hypermethylation. The mRNA expression of FHIT analyzed in ALL cases associated with a reduced pattern of expression. Significantly decreased expression was observed in majority of ALL cases (75%) than healthy controls (p = 0.01). The findings of the current study with FHIT mRNA down regulation clearly demonstrated the plausible involvement of FHIT gene expression to play an important role in the pathogenesis of ALL development. In a study by Hallas et al. [22] that confirms and agree with our report, loss of Fhit protein expression was detected in a majority of primary ALL cases and leukemia cell lines. Our results were in accordance with Chen et al. [31], where the expression of FHIT mRNA or protein was reported to be altered in 70% of cases. Further, Malak et al. [32] also agreed with our results wherein significantly less expression of FHIT mRNA was demonstrated for childhood ALL. In yet another report, Iwai et al. [21] demonstrated the RNA expression of the FHIT gene in cases of leukemia (ALL n = 11) and acute myeloid leukemia (n = 40). These results were consistent with our study and showed FHIT gene expression was totally lost or significantly reduced in 46% of the ALL cases and even 55% AML cases [33]. This also signifies the diverse role of FHIT in different types of leukemia. Reduced or loss of FHIT expression has been confirmed in solid tumors like head and neck, GIT, renal cell, breast, cervical cancers and others [34,35,36]. It has been substantiated by Siprashvili et al. [37] that transfection of FHIT into tumorigenic cell lines reduces tumor process in nude mice; pointing to the fact that FHIT acts as a tumor suppresser gene. Studies have observed that dysregulation or loss of FHIT mRNA expression occurs frequently in various cancers [38,39,40] but loss of the Fhit protein has been seen in various hematological malignancies as well [40]. Substantial evidence from a few studies suggested that the alteration of normal FHIT function could be an important event in the pathogenesis of many human hematological malignancies including ALL and the altered FHIT gene expression is specifically identified factor and highly persistent event in leukemia [21, 41]. The frequent alteration in particular reduction in FHIT expression as found in this study in ALL and other quoted studies gives an evidence that inactivating changes at the FHIT locus could supplement for the development to leukemia’s [22]. When the results of the studies are taken together with our report where clinical sample of ALL were directly examined for FHIT expression, the results demonstrate that dysregulation of FHIT expression has a plausible link in the pathogenesis of ALL. Evidences show that numerous tumor suppressor genes are inactivated by promoter region methylation in different malignancies like p15INK4B gene was frequently inactivated by promoter methylation in myelodysplastic syndrome and AML [42]. Similarly FHIT gene methylation has been depicted in few solid tumors, like GIT tumors, lung and breast cancers, where it was linked to dysregulated expression [43, 44]. Further, loss or reduced expression of FHIT protein due to its altered methylation was shown to be connected to progression of disease in certain malignancies. [45, 46] In the current study, the results showed FHIT mRNA expression was significantly decreased in those ALL cases that exhibited with hypermethylated FHIT sequences (p = 0.0008). In concordance to our results, study by Bahari et al. [47] showed that the FHIT gene hypermethylation was significantly abundant in ALL cases with FHIT expression seen significantly lower in ALL patients.
The genetic and epigenetic modification in critical regulatory genes may affect gene expression or the structure and function of specific gene products which can lead to leukemia development. In this investigation, hypermethylation of the FHIT gene was found to be considerably greater in ALL patients than in healthy controls. Our findings show that epigenetic changes and abnormal gene expression of FHIT in ALL patients may play a role in the disease’s progression. Consistent with our findings, Malak et al. [32] found a substantial decrease in the expression of FHIT in ALL compared to healthy controls. Furthermore, another study demonstrated that when compared to controls, ALL samples had significantly lower expression of FHIT with an increase in methylation frequency [31]. The association becomes crucial according to several studies as DNA methylation is the most often identified change in individuals with ALL. Further, FHIT gene deregulation in particular the decreased expression due to promoter gene methylation has been related to solid tumors for disease progression [33, 48]. In accordance to our report, a study by Hallas et al. [22] established the role of FHIT gene alteration in ALL where loss of FHIT mRNA and its product in majority of cases in contrast to their presence in almost all healthy control samples specified its plausible contribution to the pathogenesis ALL. FHIT gene as reported is emerging as a putative therapeutic target not only for ALL but other cancer types as well. A meta-analysis conducted by Wu et al. [16] where it was shown that the inactivation of FHIT gene by hypermethylation is associated with an elevated risk of non-small cell lung cancer. Therefore, the aberrant methylation patterns along with decrease in the expression of FHIT observed as seen in our study and other ones cannot be interpreted as simple secondary event, but rather must play a critical role in determining the malignant phenotype.
5 Conclusion
The current study advocates that FHIT gene hypermethylation and decreased mRNA may be linked in the pathogenesis of ALL and provide an evidence for the role of FHIT in development of ALL. However, a better interpretation of FHIT methylation and its expression may be a way forward to gain knowledge into epigenetic mechanisms of ALL to devise a strategy for treatment and unravel its different facets for chemotherapeutic outcome.
Data availability
The data supporting the results of this study are available within the paper and it’s Supplementary Information.
References
Howlader N, et al. SE9ER cancer statistics review, 1975–2017. Bethesda: National Cancer Institute; 2017.
Pandith AA, Siddiqi MA. Burden of cancers in the valley of Kashmir: 5 year epidemiological study reveals a different scenario. Tumor Biol. 2012;33:1629–37.
Sofi MA, et al. Profile of pediatric tumors: a 10-year study at a tertiary care center in North India. J Rad Cancer Res. 2022;13(3):126.
Maloney KW, et al. Outcome in children with standard-risk B-cell acute lymphoblastic leukemia: results of Children’s oncology group trial AALL0331. J Clin Oncol. 2020;38(6):602.
Schultz KR, et al. Philadelphia chromosome-negative very high-risk acute lymphoblastic leukemia in children and adolescents: results from Children’s oncology group study AALL0031. Leukemia. 2014;28(4):964–7.
Nardi V, et al. Clinical response to larotrectinib in adult Philadelphia chromosome–like ALL with cryptic ETV6-NTRK3 rearrangement. Blood Adv. 2020;4(1):106–11.
Bhatla T, et al. The biology of relapsed acute lymphoblastic leukemia: opportunities for therapeutic interventions. J Pediatr Hematol Oncol. 2014;36(6):413.
Mullighan CG, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360(5):470–80.
Tasian SK, et al. Aberrant STAT5 and PI3K/mTOR pathway signaling occurs in human CRLF2-rearranged B-precursor acute lymphoblastic leukemia. Blood, J Am Soc Hematol. 2012;120(4):833–42.
Roberts KG, et al. ETV6-NTRK3 induces aggressive acute lymphoblastic leukemia highly sensitive to selective TRK inhibition. Blood, J Am Soc Hematol. 2018;132(8):861–5.
Roberts KG, Mullighan CG. The biology of B-progenitor acute lymphoblastic leukemia. Cold Spring Harb Perspect Med. 2019;10(7): a034835.
Gomes AM, et al. Adult B-cell acute lymphoblastic leukemia cells display decreased PTEN activity and constitutive hyperactivation of PI3K/Akt pathway despite high PTEN protein levels. Haematologica. 2014;99(6):1062.
Pekarsky Y, et al. FHIT: from gene discovery to cancer treatment and prevention. Lancet Oncol. 2002;3(12):748–54.
Sard L, et al. The tumor-suppressor gene FHIT is involved in the regulation of apoptosis and in cell cycle control. Proc Natl Acad Sci. 1999;96(15):8489–92.
Sevignani C, et al. Restoration of fragile histidine triad (FHIT) expression induces apoptosis and suppresses tumorigenicity in breast cancer cell lines. Can Res. 2003;63(6):1183–7.
Wu X, et al. The clinicopathological significance and ethnic difference of FHIT hypermethylation in non-small-cell lung carcinoma: a meta-analysis and literature review. Drug Des Dev Ther. 2016;15:699–709.
Zaki SM, et al. Analysis of FHIT gene methylation in egyptian breast cancer women: association with clinicopathological features. Asian Pac J Cancer Prev. 2015;16(3):1235–9.
Yin D-T, et al. Association of the promoter methylation and protein expression of fragile histidine triad (FHIT) gene with the progression of differentiated thyroid carcinoma. Int J Clin Exp Pathol. 2010;3(5):482.
Kapitanovic S, et al. Reduced FHIT expression is associated with tumor progression in sporadic colon adenocarcinoma. Exp Mol Pathol. 2015;96(1):92–7.
Al-Temaimi RA, et al. Reduced FHIT expression is associated with mismatch repair deficient and high CpG island methylator phenotype colorectal cancer. J Histochem Cytochem. 2013;61:627–38.
Iwai T, et al. Frequent aberration of FHIT gene expression in acute leukemias. Can Res. 1998;58(22):5182–7.
Hallas C, et al. Loss of FHIT expression in acute lymphoblastic leukemia. Clin Cancer Res. 1999;5(9):2409–14.
Derwich K, et al. Long-term results in children with standard risk acute lymphoblastic leukaemia treated with 5.0 g/m 2 versus 3.0 g/m 2 methotrexate iv according to the modified ALL-BFM 90 protocol. The report of Polish paediatric Leukemia/lymphoma study group. Memo-Mag Eur Medl Oncol. 2011;4:184–9.
Gholam RB, et al. FHIT promoter DNA methylation and expressionanalysis in childhood acute lymphoblastic leukemia. Oncol Lett. 2017;1:5034–8.
Suh S-S, et al. FHIT suppresses epithelial-mesenchymal transition (EMT) and metastasis in lung cancer through modulation of microRNAs. PLoS Genet. 2014;10(10): e1004652.
Garnis C, Buys TPH, Lam WL. Genetic alteration and gene expression modulation during cancer progression. Mol Cancer. 2004;3(1):1–23.
Yan W, et al. The clinicopathological significance of FHIT hypermethylation in non-small cell lung cancer, a meta-analysis and literature review. Sci Rep. 2016;6(1):19303.
Dhillon VS, Shahid M, Husain SA. CpG methylation of the FHIT, FANCF, cyclin-D2, BRCA2 and RUNX3 genes in granulosa cell tumors (GCTs) of ovarian origin. Mol Cancer. 2004;3(1):1–8.
Zheng S, et al. Hypermethylation of the 5â€2 CpG island of the FHIT gene is associated with hyperdiploid and translocation-negative subtypes of pediatric leukemia. Can Res. 2004;64(6):2000–6.
Kiss DL, et al. Impact of FHIT loss on the translation of cancer-associated mRNAs. Mol Cancer. 2017;16(1):1–13.
Chen X, et al. Expression of fragilehistidine triad (FHIT) and WW-domain oxidoreductase gene(WWOX) in nasopharyngeal carcinoma. Asian Pac J Cancer. 2013;14:165–71.
Malak C, Elghanam D, Elbossaty W. FHIT gene expression in acute lymphoblastic leukemia and its clinical significance. Asian Pac J Cancer Prev. 2015;16:8197–201.
Takebayashi Y, et al. Loss of heterozygosity of nucleotide excision repair factors in sporadic ovarian, colon and lung carcinomas: implication for their roles of carcinogenesis in human solid tumors. Cancer Lett. 2001;174(2):115–25.
Xiao G-H, et al. The FHIT gene product is highly expressed in the cytoplasm of renal tubular epithelium and is down-regulated in kidney cancers. Am J Pathol. 1997;151(6):1541.
Virgilio L, et al. FHIT gene alterations in head and neck squamous cell carcinomas. Proc Natl Acad Sci. 1996;93(18):9770–5.
Baffa R, et al. Loss of FHIT expression in gastric carcinoma. Can Res. 1998;58(20):4708–14.
Siprashvili Z, et al. Replacement of Fhit in cancer cells suppresses tumorigenicity. Proc Natl Acad Sci. 1997;94(25):13771–6.
Carapeti M, et al. Aberrant transcripts of the FHIT gene are expressed in normal and leukaemic haemopoietic cells. Br J Cancer. 1998;78(5):601–5.
Luan X, Ramesh KH, Cannizzaro LA. FHIT gene transcript alterations occur frequently in myeloproliferative and myelodysplastic diseases. Cytogenet Genome Res. 1998;81(3–4):183–8.
Peters UR, et al. Aberrant FHIT mRNA transcripts are present in malignant and normal haematopoiesis, but absence of FHIT protein is restricted to leukaemia. Oncogene. 1999;18(1):79–85.
Sugimoto K, et al. Decreased or altered expression of the FHIT gene in human leukemias. Stem Cells. 1997;15(3):223–8.
Agrawal S, et al. DNA methylation of tumor suppressor genes in clinical remission predicts the relapse risk in acute myeloid leukemia. Cancer Res. 2007;67:1370–7.
Campiglio M, et al. FHIT loss of function in human primary breast cancer correlates with advanced stage of the disease. Can Res. 1999;59(16):3866–9.
Tseng JE, et al. Loss of Fhit is frequent in stage I non-small cell lung cancer and in the lungs of chronic smokers. Can Res. 1999;59(19):4798–803.
Connolly DC, et al. Loss of fhit expression in invasive cervical carcinomas and intraepithelial lesions associated with invasive disease. Clin Cancer Res. 2000;6(9):3505–10.
Guler G, Uner A, Guler N. Concordant loss of fragile gene expression early in breast cancer development. Pathol Int. 2005;55:471–8.
Bahari G, et al. FHIT promoter DNA methylation and expression analysisin childhood acute lymphoblastic leukemia. Oncol Lett. 2017;14:5034–8.
Mady HH, Melhem MF. FHIT protein expression and its relation to apoptosis, tumor histologic grade and prognosis in colorectal adenocarcinoma: an immunohistochemical and image analysis study. Clin Exp Metas. 2002;19(4):351.
Acknowledgements
The current studyacknowledges SERB DST Govt. of India (EMR/2014/001089 dated 24/05/2018) for funding and thank them for their financial support. All the patients are duly acknowledged for their consent to participate in the study.
Funding
DST, SERB, EMR/2014/001089.
Author information
Authors and Affiliations
Contributions
AAP, designed the study, analyzed the data and drafted the manuscript. FM participated to the study design, collected samples and patient information and performed experiments. FG, SG, SN, JR provided samples and patient related details. IQ, SMB helped in experimentation and FAG in statistical analysis. SAR, SK participated in the draft writing. All the authors accepted the manuscript format for submission.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The current study was conducted as per the Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving human samples. An informed consent was taken from each patient as per the norms of Institutional Ethics Committee before sample collection. The current study protocol was approved from Institutional Ethical Committee (IEC/SKIMS protocol: 99/2016).
Competing interests
Authors declare to have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Supplementary Figure.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mohammad, F., Pandith, A.A., Rasool, S.U.A. et al. Significance and implications of FHIT gene expression and promoter hypermethylation in acute lymphoblastic leukemia (ALL). Discov Onc 15, 108 (2024). https://doi.org/10.1007/s12672-024-00971-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-024-00971-9