Abstract
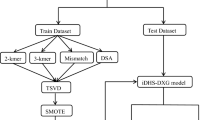
DNA N6-methyladenine (6 mA), as an essential component of epigenetic modification, cannot be neglected in genetic regulation mechanism. The efficient and accurate prediction of 6 mA sites is beneficial to the development of biological genetics. Biochemical experimental methods are considered to be time-consuming and laborious. Most of the established machine learning methods have a single dataset. Although some of them have achieved cross-species prediction, their results are not satisfactory. Therefore, we designed a novel statistical model called i6mA-VC to improve the accuracy for 6 mA sites. On the one hand, kmer and binary encoding are applied to extract features, and then gradient boosting decision tree (GBDT) embedded method is applied as the feature selection strategy. On the other hand, DNA sequences are represented by vectors through the feature extraction method of ring-function-hydrogen-chemical properties (RFHCP) and the feature selection strategy of ExtraTree. After fusing the two optimal features, a voting classifier based on gradient boosting decision tree (GBDT), light gradient boosting machine (LightGBM) and multilayer perceptron classifier (MLPC) is constructed for final classification and prediction. The accuracy of Rice dataset and M.musculus dataset with five-fold cross-validation are 0.888 and 0.967, respectively. The cross-species dataset is selected as independent testing dataset, and the accuracy reaches 0.848. Through rigorous experiments, it is demonstrated that the proposed predictor is convincing and applicable. The development of i6mA-VC predictor will become an effective way for the recognition of N6-methyladenine sites, and it will also be beneficial for biological geneticists to further study gene expression and DNA modification. In addition, an accessible web-server for i6mA-VC is available from http://www.zhanglab.site/.







Similar content being viewed by others
References
Vanyushin BF, Tkacheva SG, Belozersky AN (1970) Rare bases in animal DNA. Nature 225:948–949. https://doi.org/10.1038/225948a0
Vanyushin BF, Belozersky AN, Kokurina NA, Kadirova DX (1968) 5-Methylcytosine and 6-Methylaminopurine in bacterial DNA. Nature 218:1066–1067. https://doi.org/10.1038/2181066a0
Dunn DB, Smith JD (1955) Occurrence of a new base in the deoxyribonucleic acid of a strain of bacterium coli. Nature 175:336–337. https://doi.org/10.1038/175336a0
Unger G, Venner H (1966) Remarks on minor bases in spermatic desoxyribonucleic acid. Hoppe Seyler Z physiol Chem 344:280–283
Campbell JL, Kleckner N (1990) E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62:967–979. https://doi.org/10.1016/0092-8674(90)90271-F
Robbins-Manke JL, Zdraveski ZZ, Marinus M, Essigmann JM (2005) Analysis of global gene expression and double-strand-break formation in DNA adenine methyltransferase- and mismatch repair-deficient Escherichia coli. J Bacteriol 187:7027–7037. https://doi.org/10.1128/JB.187.20.7027-7037.2005
Pukkila PJ, Peterson J, Herman G, Modrich P, Meselson M (1983) Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics 104:571–582. https://doi.org/10.1093/genetics/104.4.571
Luria SE, Human ML (1952) A nonhereditary, host-induced variation of bacterial viruses. J Bacteriol 64:557–569. https://doi.org/10.1007/BF00410835
Meselson M, Yuan R (1968) DNA restriction enzyme from E. coli. Nature 217:1110–1114. https://doi.org/10.1038/2171110a0
Arber W, Dussoix D (1962) Host specificity of DNA produced by Escherichia coli. J Mol Biol 5:18–36. https://doi.org/10.1016/S0022-2836(62)80058-8
Bird AP (1978) Use of restriction enzymes to study eukaryotic DNA methylation: II. The symmetry of methylated sites supports semi-conservative copying of the methylation pattern. J. Mol. Biol. 118:49–60. https://doi.org/10.1016/0022-2836(78)90242-5
Pomraning KR, Smith KM, Freitag M (2009) Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods 47:142–150. https://doi.org/10.1016/j.ymeth.2008.09.022
Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW (2010) Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 7:461–465. https://doi.org/10.1038/nmeth.1459
Krais AM, Cornelius MG, Schmeiser HH (2010) Genomic N6-methyladenine determination by MEKC with LIF. Electrophoresis 31:3548–3551. https://doi.org/10.1002/elps.201000357
Greer E, Blanco M, Gu L, Sendinc E, Liu J, Aristizabal-Corrales D, Hsu CH, Aravind L, He C, Shi Y (2015) DNA Methylation on N6-Adenine in C. elegans. Cell 161:868–878. https://doi.org/10.1016/j.cell.2015.04.005
Zhou C, Wang C, Liu H, Zhou Q, Liu Q, Guo Y, Peng T, Song J, Zhang J, Chen L, Zhao Y, Zeng Z, Zhou D-X (2018) Identification and analysis of adenine N6-methylation sites in the rice genome. Nat Plants 4:554–563. https://doi.org/10.1038/s41477-018-0214-x
Chen W, Lv H, Nie F, Lin H (2019) i6mA-Pred: identifying DNA N6-methyladenine sites in the rice genome. Bioinformatics 35:2796–2800. https://doi.org/10.1093/bioinformatics/btz015
Le NQK (2019) iN6-methylat (5-step): identifying DNA N6-methyladenine sites in rice genome using continuous bag of nucleobases via Chou’s 5-step rule. Mol Genet Genomics 294:1173–1182. https://doi.org/10.1007/s00438-019-01570-y
Feng P, Yang H, Ding H, Lin H, Chen W, Chou KC (2018) iDNA6mA-PseKNC: Identifying DNA N6-methyladenosine sites by incorporating nucleotide physicochemical properties into PseKNC. Genomics. https://doi.org/10.1016/j.ygeno.2018.01.005
Pian C, Zhang G, Li F, Fan X (2019) MM-6mAPred: identifying DNA N6-methyladenine sites based on Markov Model. Bioinformatics 36:388–392. https://doi.org/10.1093/bioinformatics/btz556
Huang Q, Zhang J, Wei L, Guo F, Zou Q (2020) 6mA-RicePred: a method for identifying DNA N6-Methyladenine sites in the rice genome based on feature fusion. Front Plant Sci 11:4. https://doi.org/10.3389/fpls.2020.00004
Kong L, Zhang L (2019) i6mA-DNCP: computational identification of DNA N6-Methyladenine sites in the rice genome using optimized dinucleotide-based features. Genes 10:828. https://doi.org/10.3390/genes10100828
Liu Z, Dong W, Jiang W, He Z (2019) csDMA: an improved bioinformatics tool for identifying DNA 6 mA modifications via Chou’s 5-step rule. Sci Rep-Uk 9:13109–13118. https://doi.org/10.1038/s41598-019-49430-4
Wahab A, Ali SD, Tayara H, Chong KT (2019) iIM-CNN: intelligent identifier of 6mA sites on different species by using convolution neural network. IEEE Access. https://doi.org/10.1109/ACCESS.2019.2958618
Tahir M, Tayara H, Chong KT (2019) iDNA6mA (5-step rule): Identification of DNA N6-methyladenine sites in the rice genome by intelligent computational model via chou’s 5-step rule. Chemometr Intell Lab 189:96–101. https://doi.org/10.1016/j.chemolab.2019.04.007
Park S, Wahab A, Nazari I, Ryu JH, Chong KT (2020) i6mA-DNC: Prediction of DNA N6-Methyladenosine sites in rice genome based on dinucleotide representation using deep learning. Chemometr Intell Lab 204:104102. https://doi.org/10.1016/j.chemolab.2020.104102
Hao L, Dao FY, Guan ZX, Zhang D, Lin H (2019) iDNA6mA-Rice: a computational tool for detecting n6-methyladenine sites in rice. Front Genet 10:793. https://doi.org/10.3389/fgene.2019.00793
Basith S, Manavalan B, Shin TH, Lee G (2019) SDM6A: A web-based integrative machine-learning framework for predicting 6mA sites in the rice genome. Mol Ther-Nucl Acids. https://doi.org/10.1016/j.omtn.2019.08.011
Liu W, Li H (2020) SICD6mA: identifying 6ma sites using deep memory network. BioRxiv. https://doi.org/10.1101/2020.02.02.930776
Yu H, Dai Z (2019) SNNRice6mA: a deep learning method for predicting DNA N6-methyladenine sites in rice genome. Front Genet 10:1071–1077. https://doi.org/10.3389/fgene.2019.01071
Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. https://doi.org/10.1093/bioinformatics/bts565
Liu B, Liu F, Wang X, Chen J, Fang L, Chou KC (2015) Pse-in-One: a web server for generating various modes of pseudo components of DNA, RNA, and protein sequences. Nucleic Acids Res. https://doi.org/10.1093/nar/gkv458
Liu B, Wu H, Chou KC (2017) Pse-in-One 20: an improved package of web servers for generating various modes of pseudo components of DNA, RNA, and protein sequences. Nat Sci 9:67–91. https://doi.org/10.4236/ns.2017.94007
Chen Z, Zhao P, Li F, Marquez-Lago TT, Leier A, Revote J, Zhu Y, Powell DR, Akutsu T, Webb GI, Chou KC, Smith AI, Daly RJ, Li J, Song J (2019) iLearn: an integrated platform and meta-learner for feature engineering, machine learning analysis and modeling of DNA, RNA and protein sequence data. Brief Bioinform. https://doi.org/10.1093/bib/bbz041
Rafsanjani M, Sajid A, Dewan MF, Swakkhar S, Alok S, Abdollah D (2019) PyFeat: a Python-based effective feature generation tool for DNA RNA and protein sequences. Bioinformatics 35:3831–3833. https://doi.org/10.1093/bioinformatics/btz165
Chen Z, Zhao P, Li F, Leier A, Marquez-Lago TT, Wang Y, Webb GI, Smith AI, Daly RJ, Chou KC, Song J (2018) iFeature: a python package and web server for features extraction and selection from protein and peptide sequences. Bioinformatics 34:2499–2502. https://doi.org/10.1093/bioinformatics/bty140
He J, Fang T, Zhang Z, Huang B, Zhu X, Xiong Y (2018) PseUI: Pseudouridine sites identification based on RNA sequence information. BMC Bioinformatics 19:306. https://doi.org/10.1186/s12859-018-2321-0
Su ZD, Huang Y, Zhang ZY, Zhao YW, Wang D, Chen W, Chou KC, Lin H (2018) iLoc-lncRNA: predict the subcellular location of lncRNAs by incorporating octamer composition into general PseKNC. Bioinformatics. https://doi.org/10.1093/bioinformatics/bty508
Wang H, Ding Y, Tang J, Zou Q, Guo F (2021) Identify RNA-associated subcellular localizations based on multi-label learning using Chou’s 5-steps rule. BMC Genomics 22:1–14. https://doi.org/10.1186/s12864-020-07347-7
Zhen C, Pan X, Yang Y, Huang Y, Shen HB (2018) The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics 34:2185–2194. https://doi.org/10.1093/bioinformatics/bty085
Bari ATMG, Reaz MR, Choi HJ, Jeong BS (2013) DNA encoding for splice site prediction in large DNA sequence. Database Syst Adv Appl. https://doi.org/10.1007/978-3-642-40270-8_4
Chen W, Feng P, Tang H, Ding H, Lin H (2016) Identifying 2’-O-methylationation sites by integrating nucleotide chemical properties and nucleotide compositions. Genomics 107:255–258. https://doi.org/10.1016/j.ygeno.2016.05.003
Chen W, Yang H, Feng P, Ding H, Lin H (2017) iDNA4mC: identifying DNA N4-methylcytosine sites based on nucleotide chemical properties. Bioinformatics 33:3518–3523. https://doi.org/10.1093/bioinformatics/btx479
Wei L, Chen H, Su R (2018) M6APred-EL: a sequence-based predictor for identifying N6-methyladenosine sites using ensemble learning. Mol Ther Nucleic Acids 12:635–644. https://doi.org/10.1016/j.omtn.2018.07.004
Wei L, Su R, Luan S, Liao Z, Manavalan B, Zou Q, Shi X (2019) Iterative feature representations improve N4-methylcytosine site prediction. Bioinformatics 35:4930–4937. https://doi.org/10.1093/bioinformatics/btz408
Lv Z, Jin S, Ding H, Zou Q (2019) A random forest sub-golgi protein classifier optimized via dipeptide and amino acid composition features. Front Bioeng Biotech 7(2019):215. https://doi.org/10.3389/fbioe.2019.00215.eCollection
Fu X, Cai L, Zeng X, Zou Q (2020) StackCPPred: a stacking and pairwise energy content-based prediction of cell-penetrating peptides and their uptake efficiency. Bioinformatics 36:3028–3034. https://doi.org/10.1093/bioinformatics/btaa131
Zhang S, Qiao H (2020) KD-KLNMF: identification of lncRNAs subcellular localization with multiple features and nonnegative matrix factorization. Anal Biochem. https://doi.org/10.1016/j.ab.2020.113995
Friedman JH (2001) Greedy function approximation: a gradient boosting machine. Ann Stat 29:1189–1232. https://doi.org/10.2307/2699986
Ke G, Meng Q, Finley T, Wang T, Chen W, Ma W, Ye Q, Liu TY (2017) LightGBM: a highly efficient gradient boosting decision tree, In: 31st Conference Neural Information Processing Systems 30, pp 3149–3157. doi: https://doi.org/10.5555/3294996. 3295074.
Chou KC, Zhang CT (2008) Prediction of protein structural classes. Crit Rev Biochem Mol 30:275–349. https://doi.org/10.3109/10409239509083488
Su R, Hu J, Zou Q, Manavalan B, Wei L (2020) Empirical comparison and analysis of web-based cell-penetrating peptide prediction tools. Brief Bioinform 21:408–420. https://doi.org/10.1093/bib/bby124
Manavalan B, Basith S, Shin TH, Wei L, Lee G (2019) mAHTPred: a sequence-based meta-predictor for improving the prediction of anti-hypertensive peptides using effective feature representation. Bioinformatics 35:2757–2765. https://doi.org/10.1093/bioinformatics/bty1047
Jia J, Liu Z, Xiao X, Liu B, Chou KC (2015) iPPI-Esml: An ensemble classifier for identifying the interactions of proteins by incorporating their physicochemical properties and wavelet transforms into PseAAC. J Theor Biol 377:47–56. https://doi.org/10.1016/j.jtbi.2015.04.011
Basith S, Manavalan B, Shin TH, Lee G (2018) iGHBP: Computational identification of growth hormone binding proteins from sequences using extremely randomised tree. Comput Struct Biotec 16:412–420. https://doi.org/10.1016/j.csbj.2018.10.007
Manavalan B, Govindaraj RG, Shin TH, Kim MO, Lee G (2018) iBCE-EL: a new ensemble learning framework for improved linear B-cell epitope prediction. Front Immunol 9:1695. https://doi.org/10.3389/fimmu.2018.01695
Wei L, Luan S, Nagai LAE, Su R, Zou Q (2019) Exploring sequence-based features for the improved prediction of DNA N4-methylcytosine sites in multiple species. Bioinformatics 35:1326–1333. https://doi.org/10.1093/bioinformatics/bty824
Meng C, Guo F, Zou Q (2020) CWLy-SVM: a support vector Machine-based tool for identifying cell wall lytic enzymes. Comput Biol Chem 87:107304. https://doi.org/10.1016/j.compbiolchem.2020.107304
Zhang S, Zhu F, Yu Q, Zhu X (2021) Identifying DNA-binding proteins based on multi-features and LASSO feature selection. Biopolymers. https://doi.org/10.1002/bip.23419
Crooks GE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190. https://doi.org/10.1101/gr.849004
He W, Jia C, Zou Q (2018) 4mCPred: machine learning methods for DNA N4-methylcytosine sites prediction. Bioinformatics 35:593–601. https://doi.org/10.1093/bioinformatics/bty668
Wang J, Zhang S (2021) PA-PseU: an incremental passive-aggressive based method for identifying RNA pseudouridine sites via Chou’s 5-steps rule. Chemometr Intell Lab. https://doi.org/10.1016/j.chemolab.2021.104250
Li J, Pu Y, Tang J, Zou Q, Guo F (2020) DeepATT: a hybrid category attention neural network for identifying functional effects of DNA sequences. Brief Bioinform. https://doi.org/10.1093/bib/bbaa159
He S, Guo F, Zou Q, Ding H (2020) MRMD2.0: a python tool for machine learning with feature ranking and reduction. Curr. Bioinform. 15:1213–1221. https://doi.org/10.2174/1574893615999200503030350
Zhang YP, Zou Q (2020) PPTPP: a novel therapeutic peptide prediction method using physicochemical property encoding and adaptive feature representation learning. Bioinformatics 36:3982–3987. https://doi.org/10.1093/bioinformatics/btaa275
Zou H, Hastie T (2005) Regularization and variable selection via the elastic net. J R Stat Soc 67:768–768. https://doi.org/10.1111/j.1467-9868.2005.00527.x
Breiman L (2001) Random forest. Mach Learn 45:5–32. https://doi.org/10.1023/A:1010933404324
Vapnik VN (1998) Statistical learning theory. In: New York: Wiley, p 1–768. doi: https://doi.org/10.1007/978-1-4419-1428-6_5864.
Chen T, Guestrin C (2016) XGBoost: A Scalable Tree Boosting System. Acm sigkdd international conference on knowledge discovery and data mining, p 785–794 doi: https://doi.org/10.1145/2939672.2939785.
Geurts P, Ernst D, Wehenkel L (2006) Extremely randomized trees. Mach Learn 63:3–42. https://doi.org/10.1007/s10994-006-6226-1
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No.11601407), the Natural Science Basic Research Program of Shaanxi (No. 2021JM-115), and the Fundamental Research Funds for the Central Universities (No. JB210715).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Xue, T., Zhang, S. & Qiao, H. i6mA-VC: A Multi-Classifier Voting Method for the Computational Identification of DNA N6-methyladenine Sites. Interdiscip Sci Comput Life Sci 13, 413–425 (2021). https://doi.org/10.1007/s12539-021-00429-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-021-00429-4