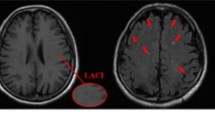
Abstract
White matter magnetic resonance hyperintensities of presumed vascular origin, which could be widely observed in elderly people, and has significant importance in multiple neurological studies. Quantitative measurement usually relies heavily on manual or semi-automatic delineation and intuitive localization, which is time-consuming and observer-dependent. Current automatic quantification methods focus mainly on the segmentation, but the spatial distribution of lesions plays a vital role in clinical diagnosis. In this study, we implemented four segmentation algorithms and compared the performances quantitatively and qualitatively on two open-access datasets. The location-specific analysis was conducted sequentially on 213 clinical patients with cerebral ischemia and lacune. The experimental results suggest that our deep-learning-based model has the potential to be integrated into the clinical workflow.





Similar content being viewed by others
References
Wardlaw JM, Smith EE, Biessels GJ, Cordonnier C, Fazekas F, Frayne R, Lindley RI, O’Brien JT, Barkhof F, Benavente OR et al (2013) Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol 12(8):822–838
Pantoni L (2010) Cerebral small vessel disease: from pathogenesis and clinical characteristics to therapeutic challenges. Lancet Neurol 9(7):689–701
Ge Y (2006) Multiple sclerosis: the role of MR imaging. Am J Neuroradiol 27(6):1165–1176
Kempton MJ, Geddes JR, Ettinger U, Williams SCR, Grasby PM (2008) Meta-analysis, database, and metaregression of 98 structural imaging studies in bipolar disorder. Arch Gen Psychiatry 65(9):1017–1032
Theodoridou A, Settas L (2006) Demyelination in rheumatic diseases. J Neurol Neurosurg Psychiatry 77(3):290–295
Hachinski V, Iadecola C, Petersen RC, Breteler MM, Nyenhuis DL, Black SE, Powers WJ, DeCarli C, Merino JG, Kalaria RN et al (2006) National institute of neurological disorders and stroke—Canadian stroke network vascular cognitive impairment harmonization standards. Stroke 37(9):2220–2241
Schmidt P (2017) Bayesian inference for structured additive regression models for large-scale problems with applications to medical imaging. Ph.D. thesis, lMU
Schmidt P, Gaser C, Arsic M, Buck D, Förschler A, Berthele A, Hoshi M, Ilg R, Schmid VJ, Zimmer C et al (2012) An automated tool for detection of flair-hyperintense white-matter lesions in multiple sclerosis. Neuroimage 59(4):3774–3783
Ithapu V, Singh V, Lindner C, Austin BP, Hinrichs C, Carlsson CM, Bendlin BB, Johnson SC (2014) Extracting and summarizing white matter hyperintensities using supervised segmentation methods in Alzheimer’s disease risk and aging studies. Hum Brain Mapp 35(8):4219–4235
Wu D, Albert M, Soldan A, Pettigrew C, Oishi K, Tomogane Y, Ye C, Ma T, Miller MI, Mori S (2019) Multi-atlas based detection and localization (MADL) for location-dependent quantification of white matter hyperintensities. Neuroimage Clin 22:101772
Lesjak Ž, Galimzianova A, Koren A, Lukin M, Pernuš F, Likar B, Špiclin Ž (2018) A novel public MR image dataset of multiple sclerosis patients with lesion segmentations based on multi-rater consensus. Neuroinformatics 16(1):51–63
Rachmadi MF, Vald’es-Hern’andez MC, Agan MLF, Perri C, Komura T, Alzheimer’s Disease Neuroimaging Initiative et al (2018) Segmentation of white matter hyperintensities using convolutional neural networks with global spatial information in routine clinical brain MRI with none or mild vascular pathology. Comput Med Imaging Gr 66:28-43
Clifford JR, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, Whitwell JL, Ward C et al (2008) The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imaging 27(4):685–691
Kuijf HJ, Biesbroek JM, de Bresser J, Heinen R, Andermatt S, Bento M, Berseth M, Belyaev M, Cardoso MJ, Casamitjana A et al (2019) Standardized assessment of automatic segmentation of white matter hyperintensities; results of the WMH segmentation challenge. IEEE Trans Med Imaging 38(11):2556–2568
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computerassisted intervention. Springer, pp 234–241
Li H, Jiang G, Zhang J, Wang R, Wang Z, Zheng W-S, Menze B (2018) Fully convolutional network ensembles for white matter hyperintensities segmentation in mr images. Neuroimage 183:650–665
Guerrero R, Qin C, Oktay O, Bowles C, Chen L, Joules R, Wolz R, Vald’es-Hernández MC, Dickie DA, Wardlaw J et al (2018) White matter hyperintensity and stroke lesion segmentation and differentiation using convolutional neural networks. Neuroimage Clin 17:918–934
He K, Zhang X, Ren S, Jian S (2016) Deep residual learning for image recognition. In: The IEEE conference on computer vision and pattern recognition (CVPR)
Kim KW, MacFall JR, Payne ME (2008) Classification of white matter lesions on magnetic resonance imaging in elderly persons. Biol Psychiatry 64(4):273–280
Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B (2017) Efficient multi-scale 3d CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 36:61–78
Milletari F, Navab N, Ahmadi S-A (2016) Vnet: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 fourth international conference on 3D vision (3DV). IEEE, pp 565–571
Hammers A, Allom R, Koepp MJ, Free SL, Myers R, Lemieux L, Mitchell TN, Brooks DJ, Duncan JS (2003) Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp 9(4):224–247
Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC (2010) N4itk: improved n3 bias correction. IEEE Trans Med Imaging 29(6):1310
Collignon A, Maes F, Delaere D, Vandermeulen D, Suetens P, Marchal G (1995) Automated multimodality image registration based on information theory. Inf Process Med Imaging 3:263–274
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, pp 1097-1105
Zhang Z, Liu Q, Wang Y (2018) Road extraction by deep residual u-net. IEEE Geosci Remote Sens Lett 15(5):749–753
Zhao L, Biesbroek JM, Shi L, Liu W, Kuijf HJ, Chu WWC, Jill MA, Lee RKL, Leung TWH, Lau AYL et al (2018) Strategic infarct location for post-stroke cognitive impairment: a multivariate lesion-symptom mapping study. J Cereb Blood Flow Metab 38(8):1299–1311
Acknowledgements
This manuscript has been accepted for presentation in The Fourth CCF Bioinformatics Conference (CBC 2019).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Jiang, W., Lin, F., Zhang, J. et al. Deep-Learning-Based Segmentation and Localization of White Matter Hyperintensities on Magnetic Resonance Images. Interdiscip Sci Comput Life Sci 12, 438–446 (2020). https://doi.org/10.1007/s12539-020-00398-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-020-00398-0