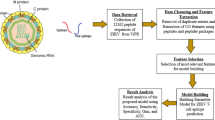
Abstract
Development of an effective machine-learning model for T-cell Mycobacterium tuberculosis (M. tuberculosis) epitopes is beneficial for saving biologist’s time and effort for identifying epitope in a targeted antigen. Existing NetMHC 2.2, NetMHC 2.3, NetMHC 3.0 and NetMHC 4.0 estimate binding capacity of peptide. This is still a challenge for those servers to predict whether a given peptide is M. tuberculosis epitope or non-epitope. One of the servers, CTLpred, works in this category but it is limited to peptide length of 9-mers. Therefore, in this work direct method of predicting M. tuberculosis epitope or non-epitope has been proposed which also overcomes the limitations of above servers. The proposed method is able to work with variable length epitopes having size even greater than 9-mers. Identification of T-cell or B-cell epitopes in the targeted antigen is the main goal in designing epitope-based vaccine, immune-diagnostic tests and antibody production. Therefore, it is important to introduce a reliable system which may help in the diagnosis of M. tuberculosis. In the present study, computational intelligence methods are used to classify T-cell M. tuberculosis epitopes. The caret feature selection approach is used to find out the set of relevant features. The ensemble model is designed by combining three models and is used to predict M. tuberculosis epitopes of variable length (7–40-mers). The proposed ensemble model achieves 82.0% accuracy, 0.89 specificity, 0.77 sensitivity with repeated k-fold cross-validation having average accuracy of 80.61%. The proposed ensemble model has been validated and compared with NetMHC 2.3, NetMHC 4.0 servers and CTLpred T-cell prediction server.






Similar content being viewed by others
References
Organization World Health (2016) Global tuberculosis report 2016. WHO. https://bit.ly/2qlTZ4j
Shah P, Mistry J, Reche PA, Gatherer D, Flower DR (2018) In silico design of mycobacterium tuberculosis epitope ensemble vaccines. Mol Immunol 97:56–62
Ferraz J, Melo F, Albuquerque MdFPM, Montenegro S, Abath F (2006) Immune factors and immunoregulation in tuberculosis. Braz J Med Biol Res 39(11):1387–1397
Flynn JL (2004) Immunology of tuberculosis and implications in vaccine development. Tuberculosis 84(1):93–101
Zhao Y, Pinilla C, Valmori D, Martin R, Simon R (2003) Application of support vector machines for T-cell epitopes prediction. Bioinformatics 19(15):1978–1984
Brusic V, Bajic VB, Petrovsky N (2004) Computational methods for prediction of T-cell epitopes a framework for modelling, testing, and applications. Methods 34(4):436–443
Bhasin M, Raghava G (2004) Prediction of CTL epitopes using QM. SVM and ANN techniques. Vaccine 22(23–24):3195–3204
Nielsen M, Lundegaard C, Worning P, Lauemøller SL, Lamberth K, Buus S, Brunak S, Lund O (2003) Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 12(5):1007–1017
Dönnes P, Elofsson A (2002) Prediction of MHC class I binding peptides, using SVMHC. BMC Bioinform 3(1):25
Pellequer JL, Westhof E, Van Regenmortel MH (1993) Correlation between the location of antigenic sites and the prediction of turns in proteins. Immunol Lett 36(1):83–99
Alix AJ (1999) Predictive estimation of protein linear epitopes by using the program PEOPLE. Vaccine 18(3):311–314
Odorico M, Pellequer JL (2003) BEPITOPE: predicting the location of continuous epitopes and patterns in proteins. J Mol Recogn 16(1):20–22
Saha S, Raghava G (2004) BcePred: prediction of continuous B-cell epitopes in antigenic sequences using physico-chemical properties. In: Nicosia G, Cutello V, Bentley PJ, Timmis J (eds) Artificial immune systems. International conference on artificial immune systems, vol 3239. Springer, Berlin, Heidelberg, pp 197–204
Saha S, Raghava G (2006) Prediction of continuous B-cell epitopes in an antigen using recurrent. Neural Netw 65(1):40–48
Chen J, Liu H, Yang J, Chou KC (2007) Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids 33(3):423–428
EL-Manzalawy Y, Dobbs D, Honavar V (2008) Predicting linear B-cell epitopes using string kernels. J Mol Recogn 21(4):243–255
Yao B, Zhang L, Liang S, Zhang C (2012) SVMTriP: a method to predict antigenic epitopes using support vector machine to integrate tri-peptide similarity and propensity. PloS One 7(9):e45152
Huang JH, Wen M, Tang LJ, Xie HL, Fu L, Liang YZ, Lu HM (2014) Using random forest to classify linear B-cell epitopes based on amino acid properties and molecular features. Biochimie 103:1–6
Yao L, HUANG ZC, Meng G, PAN XM (2015) An improved method for predicting linear B-cell epitope using deep maxout networks. Biomed Environ Sci 28(6):460–463
Shen W, Cao Y, Cha L, Zhang X, Ying X, Zhang W, Ge K, Li W, Zhong L (2015) Predicting linear B-cell epitopes using amino acid anchoring pair composition. BioData mining 8(1):1
Saha S, Raghava G (2006) AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res 34(suppl 2):W202–W209
Mohabatkar H, Mohammad Beigi M, Abdolahi K, Mohsenzadeh S (2013) Prediction of allergenic proteins by means of the concept of chou’s pseudo amino acid composition and a machine learning approach. Med Chem 9(1):133–137
Gupta S, Ansari HR, Gautam A, Raghava GP (2013) Identification of B-cell epitopes in an antigen for inducing specific class of antibodies. Biol Direct 8(1):1
Khanna D, Rana PS (2017) Multilevel ensemble model for prediction of IgA and IgG antibodies. Immunol Lett 184:51–60
Fleri W, Paul S, Dhanda SK, Mahajan S, Xu X, Peters B, Sette A (2017) The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front Immunol 8:278
Dhanda SK, Vir P, Raghava GP (2013) Designing of interferon-gamma inducing MHC class-II binders. Biol Direct 8(1):30
Vizcaíno C, Restrepo-Montoya D, Rodríguez D, Niño LF, Ocampo M, Vanegas M, Reguero MT, Martínez NL, Patarroyo ME, Patarroyo MA (2010) Computational prediction and experimental assessment of secreted/surface proteins from mycobacterium tuberculosis H37Rv. PLoS Comput Biol 6(6):e1000824
Nielsen M, Lund O (2009) NN-align. an artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinform 10(1):296
Jensen KK, Andreatta M, Marcatili P, Buus S, Greenbaum JA, Yan Z, Sette A, Peters B, Nielsen M (2018) Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 154(3):394–406
Buus S, Lauemøller S, Worning P, Kesmir C, Frimurer T, Corbet S, Fomsgaard A, Hilden J, Holm A, Brunak S (2003) Sensitive quantitative predictions of peptide-mhc binding by a query by committeeartificial neural network approach. Tissue antigens 62(5):378–384
Andreatta M, Nielsen M (2015) Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 32(4):511–517
Andreatta M, Schafer-Nielsen C, Lund O, Buus S, Nielsen M (2011) Nnalign: a web-based prediction method allowing non-expert end-user discovery of sequence motifs in quantitative peptide data. PLoS One 6(11):e26781
Nielsen M, Andreatta M (2016) NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med 8(1):33
Osorio D, Rondon-Villarreal P, Torres R (2015) Peptides: a package for data mining of antimicrobial peptides. R J 7(1):4–14
Boman H (2003) Antibacterial peptides: basic facts and emerging concepts. J Intern Med 254(3):197–215
Hofmann H, Hare E, GGobi Foundation (2016) Evaluation of diversity in nucleotide libraries, version 0.2.2. https://github.com/heike/peptider
RColorBrewer S, Deng H, Deng MH (2018) Package ‘RRF’, version 1.9. https://sites.google.com/site/houtaodeng/rrf
Therneau, T., Atkinson, B., Ripley, B., Ripley, M.B.: Package rpart. https://cran.ma.ic.ac.uk/web/packages/rpart/rpart.pdf. Accessed 20 Apr 2016 (2018)
Williams CK, Engelhardt A, Cooper T, Mayer Z, Ziem A, Scrucca L, Tang Y, Candan C, Kuhn MM (2018) Package ‘caret’, version 6.0-80. https://cran.r-project.org/web/packages/caret/caret.pdf
Hothorn T, Buehlmann P, Kneib T, Schmid M, Hofner B, Sobotka F, Scheipl F, Hofner MB (2018) Package ‘mboost’, version 2.9-1. https://github.com/boost-R/mboost
Gosso A, Gosso MA (2012) Package ‘elmnn’, version 1.0. https://cran.rproject.org/web/packages/elmNN/index.html
Hastie T, Hastie MT (2018) Package ‘gam’, version 1.16. https://cran.r-project.org/web/packages/gam/gam.pdf
Ripley B, Venables W, Ripley MB (2016) Package ‘nnet’, version 7.3-12. https://cran.r-project.org/web/packages/nnet/nnet.pdf
Karatzoglou A, Smola A, Hornik K, Karatzoglou MA (2018) Package ‘kernlab’, version 0.9-27. ftp://tdf.c3sl.ufpr.br/CRAN/web/packages/kernlab/kernlab.pdf
Bauer E, Kohavi R (1999) An empirical comparison of voting classification algorithms: Bagging, boosting, and variants. Mach Learn 36(1–2):105–139
Geluk A, Van Meijgaarden KE, Franken KL, Drijfhout JW, DSouza S, Necker A, Huygen K, Ottenhoff TH (2000) Identification of major epitopes of Mycobacterium tuberculosis AG85B that are recognized by HLA-A* 0201-restricted CD8+ T cells in HLA-transgenic mice and humans. J Immunol 165(11):6463–6471
McMurry J, Sbai H, Gennaro M, Carter E, Martin W, De Groot A (2005) Analyzing Mycobacterium tuberculosis proteomes for candidate vaccine epitopes. Tuberculosis 85(1):95–105
Lata S, Bhasin M, Raghava GP (2009) MHCBN 4.0: a database of MHC/TAP binding peptides and T-cell epitopes. BMC Res Notes 2(1):61
Nielsen M, Lundegaard C, Worning P, Hvid CS, Lamberth K, Buus S, Brunak S, Lund O (2004) Improved prediction of MHC class I and class II epitopes using a novel gibbs sampling approach. Bioinformatics 20(9):1388–1397
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khanna, D., Rana, P.S. Ensemble Technique for Prediction of T-cell Mycobacterium tuberculosis Epitopes. Interdiscip Sci Comput Life Sci 11, 611–627 (2019). https://doi.org/10.1007/s12539-018-0309-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-018-0309-0