Abstract
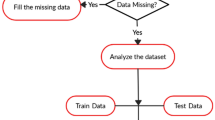
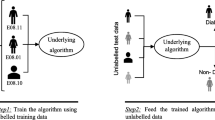
Vaccine design through experimental analysis is too costly and time taking process. By using computational intelligence approaches, cost and time can be reduced. Identification of linear B-cell epitopes is the main concern of peptide vaccine designs, immunodiagnosis, and antibody productions. It can be performed by developing a suitable machine learning model. In this paper, prediction of linear B-cell epitopes has been performed by using a bagging-based proposed ensemble model. The goal of using a bagging-based ensemble approach is to improve the prediction performance of various base classifiers. Here, five existing base classifiers are combined for ensembling to predict the B-cell epitopes or non-epitopes. This prediction is based on a bagging-based voting system, which is performed under binary class classification. The proposed ensemble model achieved 82.06% accuracy, which is better than to some existing models. Finally, the proposed ensemble model has been tested using sevenfold cross-validation, and where it provided almost consistent performance.








Similar content being viewed by others
References
Khanna D, Rana PS (2019) Improvement in prediction of antigenic epitopes using stacked generalization: an ensemble approach. IET Sys Bio 14(1):1–7. https://doi.org/10.1049/iet-syb.2018.5083
Jose L, Marta Pedro A (2017) Fundamentals and methods for T-and B-cell epitope prediction. J Immunol Res 2017:1–14. https://doi.org/10.1155/2017/2680160
Reth M, Hombach J, Wienands J, Campbell KS, Chien N, Justement LB, Cambier JC (1991) The B-cell antigen receptor complex. Immunol Today 12(6):196–201. https://doi.org/10.1016/0167-5699(91)90053-V
Saha S, Raghava GPS (2004) Bcepred: prediction of continuous b-cell epitopes in antigenic sequences using physicochemical properties. In: International Conference on Artificial Immune Systems, Catania, pp 197–204
Odorico M, Jean-Luc P (2003) Bepitope: predicting the location of continuous epitopes and patterns in proteins. J Mol Recognit 16(1):20–22. https://doi.org/10.1002/jmr.602
Pellequer JL, Westhof E, Van Regenmortel MHV (1993) Correlation between the location of antigenic sites and the prediction of turns in proteins. Immunol Lett 36(1):83–99
Alix AJP (1999) Predictive estimation of protein linear epitopes by using the program people. Vaccine 18(3):311–314
Pellequer JL, Westhof E, Van Regenmortel MH (2004) Predicting location of continuous epitopes in proteins from their primary structures. Method Enzymol 203:176–201. https://doi.org/10.1016/0076-6879(91)03010-E
Saha S, Raghava GPS (2006) Prediction of continuous b-cell epitopes in an antigen using recurrent neural network. Proteins: Structure Function Bioinformatics 65(1):40–48. https://doi.org/10.1002/prot.21078
El-Manzalawy Y, Dobbs D, Honavar V (2008) Predicting linear b-cell epitopes using string kernels. J Mol Recognit 21(4):243–255. https://doi.org/10.1002/jmr.893
Sun P, Ju H, Liu Z et al (2013) Bioinformatics resources and tools for conformational B-cell epitope prediction. Comput Math Methods Med 13(1):943636–943636
Greenbaum JA, Andersen PH, Blythe M et al (2007) Towards a consensus on datasets and evaluation metrics for developing b-cell epitope prediction tools. J Mol Recognit 20(2):75–82. https://doi.org/10.1002/jmr.815
Yao B, Zheng D, Liang S et al (2013) Conformational B-cell epitope prediction on antigen protein structures: a review of current algorithms and comparison with common binding site prediction methods. PLoS ONE 8(4):e62249. https://doi.org/10.1371/journal.pone.0062249
Luscombe NM, Greenbaum D, Gerstein M (2001) What is bioinformatics? A proposed definition and overview of the field. Method Inform Med 40(4):346–358
Larranaga P, Calvo B, Santana R et al (2006) Machine learning in bioinformatics. Brief Bioinf 7(1):86–112
Chen J, Liu H, Yang J, Chou KC (2007) Prediction of linear bcell epitopes using amino acid pair antigenicity scale. Amino Acids 33(3):423–428. https://doi.org/10.1007/s00726-006-0485-9
Yao B, Zhang L, Liang S, Zhang C (2012) SVMTrip: a method to predict antigenic epitopes using support vector machine to integrate tri-peptide similarity and propensity. PLoS ONE 7(9):e45152. https://doi.org/10.1371/journal.pone.0045152
Wee LJK, Simarmata D, Kam YW, Lisa FP, Tong JC (2010) SVM-based prediction of linear B-cell epitopes using bayes feature extraction. BMC Genomics 11(4):S21
Lin SY, Cheng CW, Chia-Yu SE (2013) Prediction of B-cell epitopes using evolutionary information and propensity scales. BMC Bioinf 14(2):S10
Wang Y, Wu W, Nicolas N, Negre NN et al (2011) Determinants of antigenicity and specificity in immune response for protein sequences. BMC Bioinf 12(1):251. https://doi.org/10.1186/1471-2105-12-251
Haung JH, Wen M, Li-Juan T, Hua-Lin X et al (2014) Using random forest to classify linear b-cell epitopes based on amino acid properties and molecular features. Biochimie 103:1–6. https://doi.org/10.1016/j.biochi.2014.03.016
Gupta VK, Rana PS (2021) Ensemble technique for toxicity prediction of small drug molecules of the antioxidant response element signaling pathway. Comput J. https://doi.org/10.1093/comjnl/bxaa001
Singh H, Ansari HR, Raghava GPS (2013) Improved method for linear B-cell epitope prediction using antigen’s primary sequence. PLoS ONE. https://doi.org/10.1371/journal.pone.0062216
Singh H, Ansari HR, Raghava GPS (2020) Lbtope: linear b-cell epitope prediction server. http://crdd.osdd.net/raghava/lbtope/data.php. Accessed 06 Dec 2020
Gupta VK, Rana PS (2019) Toxicity prediction of small drug molecules of androgen receptor using multilevel ensemble model. J Bioinf Comput Biol 17(5):1950033. https://doi.org/10.1142/S0219720019500331
Khanna D, Rana PS (2019) Ensemble technique for prediction of T-cell mycobacterium tuberculosis epitopes. Interdiscip Sci 11(4):611–627. https://doi.org/10.1007/s12539-018-0309-0
Terry T, Beth A, Brian R (2020) rpart, Package ‘rpart’—The R Project for Statistical Computing’, https://cran.rproject.org/web/packages/rpart/rpart.pdf. Accessed 12 Oct 2020
Andy L, Matthew W (2021) randomForest, Package ‘random-Forest’ - The R Project for Statistical Computing. https://cran.r-project.org/web/packages/randomForest/randomForest.pdf. Accessed 18 Oct 2020
Brian R, William V (2020) Package ‘nnet’- R Project for Statistical Computing. https://cran.rproject.org/web/packages/nnet/nnet.pdf. Accessed 12 Oct 2021
Mark C, Kjell J, George M (2020) Ada, The R Package ‘Ada’ for Stochastic Boosting. https://cran.rproject.org/web/packages/ada/ada.pdf. Accessed 15 Sept 2020
David M, Evgenia D (2019) e1071, The R Package for ‘support vector machine’. https://cran.r-project.org/web/packages/e1071/e1071.pdf. Accessed 12 Nov 2019
Gupta VK, Rana PS (2019) Activity assessment of small drug molecules in estrogen receptor using multilevel prediction model. IET sys bio. 13(3):147–158. https://doi.org/10.1049/iet-syb.2018.5068
Gupta VK, Rana PS (2020) Toxicity prediction of pre-clinical trial drugs using physicochemical properties and computational intelligence approaches. Dissertation, Thapar Institute of Engineering and Technology, Patiala
Han J, Pei J, Kamber M (2011) Data mining: concepts and techniques, 3rd edn. Elsevier, USA
Gupta VK, Rana PS (2021) Toxicity prediction of small drug molecules of aryl hydrocarbon receptor using a proposed ensemble model. Turk J Electr Eng Co 24(4):2833–2849. https://doi.org/10.3906/elk-1809-9
Achuthsankar SN, Aswathi B (2020) ‘Sensitivity, specificity, accuracy and the relationship between them’, http://www.lifenscience.com/bioinformatics/sensitivity-specificity-accuracy-and. Accessed 7 Jan 2020
Acknowledgements
We are very much thankful to the LBtope server for making the data, which is available to the public. We are also thankful for the various web servers, which have been developed to predict B-cell epitopes or non-epitopes in an antigenic sequence.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declared no conflicts of interest with respect to the authorship, research, and publication of this manuscript.
Rights and permissions
About this article
Cite this article
Gupta, V.K., Gupta, A., Jain, P. et al. Linear B-cell epitopes prediction using bagging based proposed ensemble model. Int. j. inf. tecnol. 14, 3517–3526 (2022). https://doi.org/10.1007/s41870-022-00951-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41870-022-00951-8