Abstract
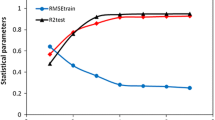
Flavonoids exhibit a high affinity for the purified cytosolic NBD (C-terminal nucleotide-binding domain) of P-glycoprotein (P-gp). To explore the affinity of flavonoids for P-gp, quantitative structure–activity relationship (QSAR) models were developed using support vector machines (SVMs). A novel method coupling a modified particle swarm optimization algorithm with random mutation strategy and a genetic algorithm coupled with SVM was proposed to simultaneously optimize the kernel parameters of SVM and determine the subset of optimized features for the first time. Using DRAGON descriptors to represent compounds for QSAR, three subsets (training, prediction and external validation set) derived from the dataset were employed to investigate QSAR. With excluding of the outlier, the correlation coefficient (R2) of the whole training set (training and prediction) was 0.924, and the R2 of the external validation set was 0.941. The root-mean-square error (RMSE) of the whole training set was 0.0588; the RMSE of the cross-validation of the external validation set was 0.0443. The mean Q2 value of leave-many-out cross-validation was 0.824. With more informations from results of randomization analysis and applicability domain, the proposed model is of good predictive ability, stability.




Similar content being viewed by others
References
Adeloye AJ, Rustum R (2012) Self-organising map rainfall–runoff multivariate modelling for runoff reconstruction in inadequately gauged basins. Hydrol Res 43:603–617
Andrews PS (2006) An investigation into mutation operators for particle swarm optimization. In: IEEE congress on evolutionary computation, 2006. CEC 2006. IEEE, p 1044–1051
Ballabio D, Vasighi M, Consonni V, Kompany-Zareh M (2011) Genetic algorithms for architecture optimisation of counter-propagation artificial neural networks. Chemom Intell Lab Syst 105:56–64
Benfenati E, Piclin N, Roncaglioni A, Vari M (2001) Factors influencing predictive models for toxicology. SAR QSAR Environ Res 12:593–603
Bernard P, Pintore M, Berthon JY, Chretien JR (2001) A molecular modeling and 3D QSAR study of a large series of indole inhibitors of human non-pancreatic secretory phospholipase A2. Eur J Med Chem 36:1–19
Boccard J, Bajot F, Di Pietro A, Rudaz S, Boumendjel A, Nicolle E, Carrupt PA (2009) A 3D linear solvation energy model to quantify the affinity of flavonoid derivatives toward P-glycoprotein. Eur J Pharm Sci 36:254–264
Boumendjel A, Bois F, Beney C, Mariotte AM, Conseil G, Di Pietro A (2001) B-ring substituted 5,7-dihydroxyflavonols with high-affinity binding to P-glycoprotein responsible for cell multidrug resistance. Bioorg Med Chem Lett 11:75–77
Boumendjel A, Beney C, Deka N, Mariotte AM, Lawson MA, Trompier D, Baubichon-Cortay H, Di Pietro A (2002a) 4-Hydroxy-6-methoxyaurones with high-affinity binding to cytosolic domain of P-glycoprotein. Chem Pharm Bull (Tokyo) 50:854–856
Boumendjel A, Di Pietro A, Dumontet C, Barron D (2002b) Recent advances in the discovery of flavonoids and analogs with high-affinity binding to P-glycoprotein responsible for cancer cell multidrug resistance. Med Res Rev 22:512–529
Caballero J, Fernández L, Garriga M, Abreu JI, Collina S, Fernández M (2007) Proteometric study of ghrelin receptor function variations upon mutations using amino acid sequence autocorrelation vectors and genetic algorithm-based least square support vector machines. J Mol Graph Model 26:166–178
Chang C-C, Lin C-J (2006) LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2:27
Chen C, Zhou X, Tian Y, Zou X, Cai P (2006) Predicting protein structural class with pseudo-amino acid composition and support vector machine fusion network. Anal Biochem 357:116–121
Conseil G, Baubichon-Cortay H, Dayan G, Jault J-M, Barron D, Di Pietro A (1998) Flavonoids: a class of modulators with bifunctional interactions at vicinal ATP-and steroid-binding sites on mouse P-glycoprotein. Proc Natl Acad Sci USA 95:9831–9836
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20:273–297
Cottrell M, Fort J-C, Pagès G (1998) Theoretical aspects of the SOM algorithm. Neurocomputing 21:119–138
Escobar MS, Kaneko H, Funatsu K (2014) Flour concentration prediction using GAPLS and GAWLS focused on data sampling issues and applicability domain. Chemom Intell Lab Syst 137:33–46
Fatemi MH, Dorostkar F (2010) QSAR prediction of D 2 receptor antagonistic activity of 6-methoxy benzamides. Eur J Med Chem 45:4856–4862
Firouzi BB, Meymand HZ, Niknam T, Mojarrad HD (2011) A novel multi-objective chaotic crazy PSO algorithm for optimal operation management of distribution network with regard to fuel cell power plants. Int J Innov Comput Inf Control 7:6395–6409
Gálvez J, Garcia-Domenech R, De J-OV, Soler R (1994) Topological approach to analgesia. J Chem Inf Comput Sci 34:1198–1203
García HL, González IM (2004) Self-organizing map and clustering for wastewater treatment monitoring. Eng Appl Artif Intell 17:215–225
Gevrey M, Comte L, De Zwart D, De Deckere E, Lek S (2010) Modeling the chemical and toxic water status of the Scheldt Basin (Belgium), using aquatic invertebrate assemblages and an advanced modeling method. Environ Pollut 158:3209–3218
Golbraikh A, Tropsha A (2000) Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection. Mol Divers 5:231–243
Golbraikh A, Tropsha A (2002) Beware of q2! J Mol Graph Model 20:269–276
Gramatica P (2006) WHIM descriptors of shape. QSAR Comb Sci 25:327–332
Habibi-Yangjeh A (2009) QSAR study of the 5-HT1A receptor affinities of arylpiperazines using a genetic algorithm–artificial neural network model. Chem Mon 140:523–530
Hamadache M, Benkortbi O, Hanini S, Amrane A, Khaouane L, Moussa CS (2015) A quantitative structure activity relationship for acute oral toxicity of pesticides on rats: validation, domain of application and prediction. J Hazard Mater 303:28–40
Hao M, Li Y, Wang Y, Zhang S (2011) Prediction of P2Y 12 antagonists using a novel genetic algorithm-support vector machine coupled approach. Anal Chim Acta 690:53–63
Helguera AM, Pérez MC, González MP (2006) A radial-distribution-function approach for predicting rodent carcinogenicity. J Mol Model 12:769–780
Isfort RJ, Wang F, Tscheiner M, Donnelly E, Bauer MB, Lefever F, Hinkle RT, Mazur AW (2005) Discovery of corticotropin releasing factor 2 receptor selective sauvagine analogues for treatment of skeletal muscle atrophy. J Med Chem 48:262–265
Khajeh A, Modarress H, Zeinoddini-Meymand H (2013) Modified particle swarm optimization method for variable selection in QSAR/QSPR studies. Struct Chem 24:1401–1409
Kohonen T, Schroeder M, Huang T (2001) Maps self-organizing. Springer, New York
Kothandan G, Gadhe CG, Madhavan T, Choi CH, Cho SJ (2011) Docking and 3D-QSAR (quantitative structure activity relationship) studies of flavones, the potent inhibitors of p-glycoprotein targeting the nucleotide binding domain. Eur J Med Chem 46:4078–4088
Lavalle SM, Branicky MS (2004) On the relationship between classical grid search and probabilistic roadmaps. Int J Robot Res 23:673–692
Li ZC, Zhou XB, Lin YR, Zou XY (2008) Prediction of protein structure class by coupling improved genetic algorithm and support vector machine. Amino Acids 35:581–590
Liu H-X, Zhang R-S, Yao X-J, Liu M-C, Hu Z-D, Fan B-T (2004) Prediction of electrophoretic mobility of substituted aromatic acids in different aqueous–alcoholic solvents by capillary zone electrophoresis based on support vector machine. Anal Chim Acta 525:31–41
Loukas YL (2001) Adaptive neuro-fuzzy inference system: an instant and architecture-free predictor for improved QSAR studies. J Med Chem 44:2772–2783
Niculescu SP (2003) Artificial neural networks and genetic algorithms in QSAR. J Mol Struct THEOCHEM 622:71–83
Pratim Roy P, Paul S, Mitra I, Roy K (2009) On two novel parameters for validation of predictive QSAR models. Molecules 14:1660–1701
Roy K, Mandal AS (2009) Predictive QSAR modeling of CCR5 antagonist piperidine derivatives using chemometric tools. J Enzym Inhib Med Chem 24:205–223
Sahigara F, Mansouri K, Ballabio D, Mauri A, Consonni V, Todeschini R (2012) Comparison of different approaches to define the applicability domain of QSAR models. Molecules 17:4791–4810
Sajan KS, Kumar V, Tyagi B (2015) Genetic algorithm based support vector machine for on-line voltage stability monitoring. Int J Electr Power Energy Syst 78:200–208
Shen J, Cui Y, Gu J, Li Y, Li L (2013) A genetic algorithm-back propagation artificial neural network model to quantify the affinity of flavonoids toward P-glycoprotein. Comb Chem High Throughput Screen 17:162–172
Shi J, Chen L, Chen W (2013) Prediction of the heat capacity for compounds based on the conjugate gradient and support vector machine methods. J Chemom 27:251–259
Shieh S-L, Liao I-E (2012) A new approach for data clustering and visualization using self-organizing maps. Expert Syst Appl 39:11924–11933
Soltani S, Abolhasani H, Zarghi A, Jouyban A (2010) QSAR analysis of diaryl COX-2 inhibitors: comparison of feature selection and train-test data selection methods. Eur J Med Chem 45:2753–2760
Tchamo DN, Dijoux-Franca MG, Mariotte AM, Tsamo E, Daskiewicz JB, Bayet C, Barron D, Conseil G, Di Pietro A (2000) Prenylated xanthones as potential P-glycoprotein modulators. Bioorg Med Chem Lett 10:1343–1345
Todeschini R, Consonni V (2000) Handbook of molecular descriptors. Wiley-VCH, Weinheim
Wang Y-H, Li Y, Yang S-L, Yang L (2005) An in silico approach for screening flavonoids as P-glycoprotein inhibitors based on a Bayesian-regularized neural network. J Comput Aided Mol Des 19:137–147
Wang X, Sun Y, Wu L, Gu S, Liu R, Liu L, Liu X, Xu J (2014) Quantitative structure–affinity relationship study of azo dyes for cellulose fibers by multiple linear regression and artificial neural network. Chemom Intell Lab Syst 134:1–9
Weaver S, Gleeson MP (2008) The importance of the domain of applicability in QSAR modeling. J Mol Graph Model 26:1315–1326
Xu Q, Wei C, Liu R, Gu S, Xu J (2015) Quantitative structure–property relationship study of β-cyclodextrin complexation free energies of organic compounds. Chemom Intell Lab Syst 146:313–321
Yap CW, Li ZR, Chen YZ (2006) Quantitative structure pharmacokinetic relationships for drug clearance by using statistical learning methods. J Mol Graph Model 24:383–395
Zernov VV, Balakin KV, Ivaschenko AA, Savchuk NP, Pletnev IV (2003) Drug discovery using support vector machines. The case studies of drug-likeness, agrochemical-likeness, and enzyme inhibition predictions. J Chem Inf Comput Sci 43:2048–2056
Zhou X, Li Z, Dai Z, Zou X (2010) QSAR modeling of peptide biological activity by coupling support vector machine with particle swarm optimization algorithm and genetic algorithm. J Mol Graph Model 29:188–196
Acknowledgements
The authors are grateful for the financial support by the Central Laboratory Open Foundation (2015ZXKF08) from the Logistics College of Chinese People’s Armed Police Forces, China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Qinggang Chen contributed equally to this work and should be considered co-first authors.
Rights and permissions
About this article
Cite this article
Cui, Y., Chen, Q., Li, Y. et al. A new model of flavonoids affinity towards P-glycoprotein: genetic algorithm-support vector machine with features selected by a modified particle swarm optimization algorithm. Arch. Pharm. Res. 40, 214–230 (2017). https://doi.org/10.1007/s12272-016-0876-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12272-016-0876-8