Abstract
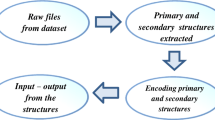
Structural class characterizes the overall folding type of a protein or its domain. Most of the existing methods for determining the structural class of a protein are based on a group of features that only possesses a kind of discriminative information for the prediction of protein structure class. However, different types of discriminative information associated with primary sequence have been completely missed, which undoubtedly has reduced the success rate of prediction. We present a novel method for the prediction of protein structure class by coupling the improved genetic algorithm (GA) with the support vector machine (SVM). This improved GA was applied to the selection of an optimized feature subset and the optimization of SVM parameters. Jackknife tests on the working datasets indicated that the prediction accuracies for the different classes were in the range of 97.8–100% with an overall accuracy of 99.5%. The results indicate that the approach has a high potential to become a useful tool in bioinformatics.




Similar content being viewed by others
References
Aguero-Chapin G, Gonzalez-Diaz H, Molina R, Varona-Santos J, Uriarte E, Gonzalez-Diaz Y (2006) Novel 2D maps and coupling numbers for protein sequences. The first QSAR study of polygalacturonases; isolation and prediction of a novel sequence from Psidium guajava L. FEBS Lett 580:723–730
Bahar I, Atilgan AR, Jernigan RL, Erman B (1997) Understanding the recognition of protein structureal classes by amino acid composition. Proteins 29:172–185
Caballero J, Fernandez L, Garriga M, Abreu JI, Collina S, Fernandez M (2007) Proteometric study of ghrelin receptor function variations upon mutations using amino acid sequence autocorrelation vectors and genetic algorithm-based least square support vector machines. J Mol Graph Model 26:166–178
Cai YD, Chou KC (2005a) Using functional domain composition to predict enzyme family classes. J Proteome Res 4:109–111
Cai YD, Chou KC (2005b) Predicting enzyme subclass by functional domain composition and pseudo amino acid composition. J Proteome Res 4:967–971
Cai YD, Chou KC (2006) Predicting membrane protein type by functional domain composition and pseudo amino acid composition. J Theor Bio 238:395–400
Cai YD, Zhou GP (2000) Prediction of protein structural classes by neural network. Biochimie 82:783–785
Cai YD, Liu XJ, Xu XB, Zhou GP (2001) Support vector machines for predicting protein structural class. BMC Bioinformatics 2:1–5
Cai YD, Liu XJ, Xu XB, Chou KC (2002) Prediction of protein structural classes by support vector machines. Comput Chem 26:293–296
Cai YD, Ricardo PW, Jen CH, Chou KC (2004) Application of SVM to predict membrane protein types. J Theor Boil 226:373–376
Cai YD, Feng KY, Lu WC, Chou KC (2006) Using logitboost classifier to predict protein structural classes. J Theor Biol 238:172–176
Cao YF, Liu S, Zhang L, Qin J, Wang J, Tang KX (2006) Prediction of protein structural class with rough sets. BMC Bioinformatics 7:1–6
Cedano J, Aloy P, P’erez-Pons JA, Querol E (1997) Relation between amino acid composition and cellular location of proteins. J Mol Biol 266:594–600
Chang CC, Lin CJ (2001) LIBSVM: a library for support vector machines. Software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm
Chen C, Tian YX, Zou XY, Cai PX, Mo JY (2006a) Using pseudo-amino acid composition and support vector machine to predict protein structural class. J Theor Biol 243:444–448
Chen C, Zhou XB, Tian YX, Zou XY, Cai PX (2006b) Predicting protein structural class with pseudo-amino acid composition and support vector machine fusion network. Anal Biochem 357:116–121
Chen J, Liu H, Yang J, Chou KC (2007) Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids 33:423–428
Chen YL, Li QZ (2007a) Prediction of apoptosis protein subcellular location using improved hybrid approach and pseudo amino acid composition. J Theor Biol 248:377–381
Chen YL, Li QZ (2007b) Prediction of the subcellular location of apoptosis proteins. J Theor Biol 245:775–783
Chou KC (1992) Energy-optimized structure of antifreeze protein and its binding mechanism. J Mol Biol 223:509–517
Chou KC (1995) A novel-approach to predicting protein structural classes in a (20–1)-D amino –acid-composition space. Proteins 21:319–344
Chou KC (1999a) Using pair-coupled amino acid composition to predict protein secondary structure content. J Protein Chem 18:473–480
Chou KC (1999b) A key driving force in determination of protein structural classes. Biochem Biophys Res Commun 264:216–224
Chou KC (2000) Prediction of protein structural classes and subcellular locations. Curr Protein Pept Sc 1:171–208
Chou KC (2001) Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins 43:246–255
Chou KC (2004) Review: structural bioinformatics and its impact to biomedical science. Curr Med Chem 11:2105–2134
Chou KC (2005) Review: progress in protein structural class prediction and its impact to bioinformatics and proteomics. Curr Protein Pept Sc 6:423–436
Chou KC, Cai YD (2002) Using functional domain composition and support vector machines for prediction of protein subcellular location. J Biol Chem 277:45765–45769
Chou KC, Cai YD (2004) Predicting protein structural class by functional domain composition. Biochem Biophys Res Commun 321:1007–1009
Chou KC, Elrod DW (1999) Protein subcellular location prediction. Protein Eng 12:107–118
Chou KC, Elrod DW (2002) Bioinformatical analysis of G-protein-coupled receptors. J Proteome Res 1:429–433
Chou KC, Elrod DW (2003) Prediction of enzyme family classes. J Proteome Res 2:183–190
Chou KC, Maggiora GM (1998) Domain structural class prediction. Protein Eng 11:523–538
Chou KC, Shen HB (2006a) Hum-PLoc: a novel ensemble classifier for predicting human protein subcellular localization. Biochem Biophys Res Commun 347:150–157
Chou KC, Shen HB (2006b) Predicting eukaryotic protein subcellular location by fusing optimized evidence-theoretic K-nearest neighbor classifiers. J Proteome Res 5:1888–1897
Chou KC, Shen HB (2007a) Euk-mPLoc: a fusion classifier for large-scale eukaryotic protein subcellular location prediction by incorporating multiple sites. J Proteome Res 6:1728–1734
Chou KC, Shen HB (2007b) Large-scale plant protein subcellular location prediction. J Cell Biochem 100:665–678
Chou KC, Shen HB (2007c) MemType-2L: a Web server for predicting membrane proteins and their types by incorporating evolution information through Pse-PSSM. Biochem Biophys Res Comm 360:339–345
Chou KC, Shen HB (2007d) Review: recent progresses in protein subcellular location prediction. Anal Biochem 370:1–16
Chou KC, Shen HB (2007e) Signal-CF: a subsite-coupled and window-fusing approach for predicting signal peptides. Biochem Biophys Res Comm 357:633–640
Chou KC, Shen HB (2008) Cell-PLoc: a package of web-servers for predicting subcellular localization of proteins in various organisms. Nat Protoc 3:153–162
Chou KC, Zhang CT (1992) A correlation-coefficient method to predicting protein-structural classes form amino-acid compositions. Eur J Biochem 207:429–433
Chou KC, Zhang CT (1994) Predicting protein folding types by distance functions that make allowances for amino acid interactions. J Biol Chem 269:22014–22020
Chou KC, Zhang CT (1995) Review: prediction of protein structural classes. Crit Rev Biochem Mol Biol 30:275–349
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20:273–297
Diao Y, Li M, Feng Z, Yin J, Pan Y (2007) The community structure of human cellular signaling network. J Theor Biol 247:608–615
Diao Y, Ma D, Wen Z, Yin J, Xiang J, Li M (2008) Using pseudo amino acid composition to predict transmembrane regions in protein: cellular automata and Lempel-Ziv complexity. Amino Acids. doi:10.1007/s00726-007-0550-z
Ding CHQ, Dubchak I (2001) Multi-class protein fold recognition using support vector machines and neural networks. Bioinformatics 17:349–358
Ding YS, Zhang TL, Chou KC (2007) Prediction of protein structure classes with pseudo amino acid composition and fuzzy support vector machine network. Protein Peptide Lett 14:811–815
Du P, Li Y (2006) Prediction of protein submitochondria locations by hybridizing pseudo-amino acid composition with various physicochemical features of segmented sequence. BMC Bioinformatics 7:518
Du QS, Wei DQ, Chou KC (2003) Correlations of amino acids in proteins. Peptides 24:1863–1869
Du QS, Jiang ZQ, He WZ, Li DP, Chou KC (2006) Amino acid principal component analysis (AAPCA) and its applications in protein structural class prediction. J Biomol Struct Dyn 23:635–640
Eshelmen LJ, Schaffer JD (1991) Preventing premature convergence in genetic algorithms by preventing incest. In: Belew RK, Booker LB (eds) Proc 4th Int Conf Genetic Algorithms. Morgan Kaufmann, San Francisco, pp 115–122
Feng KY, Cai YD, Chou KC (2005) Boosting classifier for predicting protein domain structural class. Biochem Biophys Res Commun 334:213–217
Fang Y, Guo Y, Feng Y, Li M (2008) Predicting DNA-binding proteins: approached from Chou’s pseudo amino acid composition and other specific sequence features. Amino Acids. doi:10.1007/s00726-007-0568-2
Feng ZP, Zhang CT (2000) Prediction of membrane protein types based on the hydrophobic index of amino acids. J Protein Chem 19:269–275
Gao Y, Shao SH, Xiao X, Ding YS, Huang YS, Huang ZD, Chou KC (2005) Using pseudo amino acid composition to predict protein subcellular location: approached with Lyapunov index, Bessel function, and Chebyshev filter. Amino Acids 28:373–376
Gonzalez-Diaz H, Perez-Bello A, Uriarte E, Gonzalez-Diaz Y (2006) QSAR study for mycobacterial promoters with low sequence homology. Bioorg Med Chem Lett 16:547–553
Gonzalez-Diaz H, Aguero-Chapin G, Varona J, Molina R, Delogu G, Santana L, Uriarte E, Podda G (2007a) 2D-RNA-coupling numbers: a new computational chemistry approach to link secondary structure topology with biological function. J Comput Chem 28:1049–1056
Gonzalez-Diaz H, Perez-Castillo Y, Podda G, Uriarte E (2007b) Computational chemistry comparison of stable/nonstable protein mutants classification models based on 3D and topological indices. J Comput Chem 28:1990–1995
Gonzalez-Diaz H, Vilar S, Santana L, Uriarte E (2007c) Medicinal chemistry and bioinformatics—current trends in drugs discovery with networks topological indices. Curr Top Med Chem 10:1015–1029
Guo YZ, Li M, Lu M, Wen Z, Wang K, Li G, Wu J (2006) Classifying G protein-coupled receptors and nuclear receptors based on protein power spectrum from fast Fourier transform. Amino Acids 30:397–402
Handels H, Ross T, Kreusch J, Wolff HH, Pöppl SJ (1999) Feature selection for optimized skin tumor recognition using genetic algorithms. Artif Intell Med 16:283–297
Horne DS (1988) Prediction of protein helix content from an autocorrelation analysis of sequence hydrophobicities. Biopolymers 27:451–477
Hua SJ, Sum ZR (2001) A novel method of protein secondary structure prediction with high segment overlap measure: support vector machine approach. J Mol Biol 308:397–407
Huang CL, Liao HC, Chen MC (2008) Prediction model building and feature selection with support vector machines in breast cancer diagnosis. Expert Syst Appl 34:578–587
Hsu CW, Lin CJ (2002) A simple decomposition method for support vector machine. Mach Learn 46:219–314
Jahandideh S, Abdolmaleki P, Jahandideh M, Asadabadi EB (2007a) Novel two-stage hybrid neural discriminant model for predicting proteins structural classes. Biophys Chem 128:87–93
Jahandideh S, Abdolmaleki P, Jahandideh M, Hayatshahi SHS (2007b) Novel hybrid method for the evaluation of parameters contributing in determination of protein structural classes. J Theor Biol 244:275–281
Jalali-Heravi M, Kyani A (2007) Application of genetic algorithm-kernel partial least square as a novel nonlinear feature selection method: activity of carbonic anhydrase II inhibitors. Eur J Med Chem 42:649–659
Jin LX, Fang WW, Tang HW (2003) Prediction of protein structural classes by a new measure of information discrepancy. Comput Biol Chem 23:373–380
Kedarisetti KD, Kurgan L, Dick S (2006) Classifier ensembles for protein structural class prediction with varying homology. Biochem Biophys Res Commun 348:981–988
Kim JK, Raghava GPS, Bang SY, Choi S (2006) Prediction of subcellular localization of proteins using pairwise sequence alignment and support vector machine, Pattern. Recogn Lett 27:996–1001
Kohavi R, John GH (1997) Wrappers for feature subset selection. Artif Intell 97:273–324
Kurgan LA, Stach W, Ruan J (2007) Novel scales based on hydrophobicity indices for secondary protein structure. J Theor Biol 248:354–366
Kuric L (2007). The digital language of amino acids. Amino Acids 33:653–661
LaValle SM, Branicky MS (2002) On the relationship between classical grid search and probabilistic roadmaps. Int J Robot Res 23:673–692
Levitt M, Chothia C (1976) Structural patterns in globular proteins. Nature 261:552–558
Li FM, Li QZ (2007) Using pseudo amino acid composition to predict protein subnuclear location with improved hybrid approach. Amino Acids. doi:10.1007/s00726-007-0545-9
Li ZR, Lin HH, Han LY, Jiang L, Chen X, Chen YZ (2006) PROFEAT: a web server for computing structural and physicochemical features of proteins and peptides from amino acid sequence. Nucleic Acids Res 34:32–37
Lin H, Li QZ (2007a) Predicting conotoxin superfamily and family by using pseudo amino acid composition and modified Mahalanobis discriminant. Biochem Biophys Res Commun 354:548–551
Lin H, Li QZ (2007b) Using pseudo amino acid composition to predict protein structural class: approached by incorporating 400 dipeptide components. J Comput Chem 28:1463–1466
Lin Z, Pan XM (2001) Accurate prediction of protein secondary structural content. J Protein Chem 20:217–220
Liu DQ, Liu H, Shen HB, Yang J, Chou KC (2007) Predicting secretory protein signal sequence cleavage sites by fusing the marks of global alignments. Amino Acids 32:493–496
Liu H, Wang M, Chou KC (2005a) Low-frequency Fourier spectrum for predicting membrane protein types. Biochem Biophys Res Commun 336:737–739
Liu H, Yang J, Wang M, Xue L, Chou KC (2005b) Using Fourier spectrum analysis and pseudo amino acid composition for prediction of membrane protein types. Protein J 24:385–389
Luo RY, Feng ZP, Liu JK (2002) Prediction of protein structural class by amino acid and polypeptide composition. Eur J Biochem 269:4219–4225
Lv QZ, Shen GL, Yu RQ (2003) A chaotic approach to maintain the population diversity of genetic algorithm in network training. Comput Biol Chem 27:363–371
Metfessel BA, Saurugger PN, Connelly DP, Rich SS (1993) Cross-validation of protein structural class prediction using statistical clustering and neural networks. Protein Sci 2:1170–1182
Mondal S, Bhavna R, Mohan Babu R, Ramakumar S (2006) Pseudo amino acid composition and multi-class support vector machines approach for conotoxin superfamily classification. J Theor Biol 243:252–260
Mundra P, Kumar M, Kumar KK, Jayaraman VK, Kulkarni BD (2007) Using pseudo amino acid composition to predict protein subnuclear localization: approached with PSSM. Pattern Recognit Lett 28:1610–1615
Niu B, Cai YD, Lu WC, Zheng GY, Chou KC (2006) Predicting protein structural class with AdaBoost learner. Protein Peptide Lett 13:489–492
Pan YX, Zhang ZZ, Guo ZM, Feng GY, Huang ZD, He L (2003) Application of pseudo amino acid composition for predicting protein subcellular location: stochastic signal processing approach. J Protein Chem 22:395–402
Pu X, Guo J, Leung H, Lin Y (2007) Prediction of membrane protein types from sequences and position-specific scoring matrices. J Theor Biol 247:259–265
Shen HB, Chou KC (2005a) Predicting protein subnuclear location with optimized evidence-theoretic K-nearest classifier and pseudo amino acid composition. Biochem Biophys Res Comm 337:752–756
Shen HB, Chou KC (2005b) Using optimized evidence-theoretic K-nearest neighbor classifier and pseudo amino acid composition to predict membrane protein types. Biochem Biophys Res Comm 334:288–292
Shen HB, Chou KC (2006) Ensemble classifier for protein fold pattern recognition. Bioinformatics 22:1717–1722
Shen HB, Chou KC (2007a) Gpos-PLoc: an ensemble classifier for predicting subcellular localization of Gram-positive bacterial proteins. Protein Eng Des Sel 20:39–46
Shen HB, Chou KC (2007b) Hum-mPLoc: an ensemble classifier for large-scale human protein subcellular location prediction by incorporating samples with multiple sites. Biochem Biophys Res Commun 355:1006–1011
Shen HB, Chou KC (2007c) Using ensemble classifier to identify membrane protein types. Amino Acids 32:483–488
Shen HB, Chou KC (2007d) Virus-PLoc: a fusion classifier for predicting the subcellular localization of viral proteins within host and virus-infected cells. Biopolymers 85:233–240
Shen HB, Chou KC (2008) PseAAC: a flexible web-server for generating various kinds of protein pseudo amino acid composition. Anal Biochem 373:386–388
Shen HB, Yang J, Liu XJ, Chou KC (2005) Using supervised fuzzy clustering to predict protein structural classes. Biochem Biophys Res Commun 334:577–581
Shen HB, Yang J, Chou KC (2006) Fuzzy KNN for predicting membrane protein types from pseudo amino acid composition. J Theor Biol 240:9–13
Shen HB, Yang J, Chou KC (2007a) Euk-PLoc: an ensemble classifier for large-scale eukaryotic protein subcellular location prediction. Amino Acids 33:57–67
Shen Q, Shi WM, Kong W, Ye BX (2007b) A combination of modified particle swarm optimization algorithm and support vector machine for gen selection and tumor classification. Talanta 71:1679–1683
Shi JY, Zhang SW, Pan Q, Cheng YM, Xie J (2007) Prediction of protein subcellular localization by support vector machines using multi-scale energy and pseudo amino acid composition. Amino Acids 33:69–74
Shi JY, Zhang SW, Pan Q, Zhou GP (2008) Using pseudo amino acid composition to predict protein subcellular location: approached with amino acid composition distribution. Amino Acids. doi:10.1007/s00726-007-0623-z
Sivagaminathan RK, Ramakrishnan S (2007) A hybrid approach for feature subset selection using neural networks and ant colony optimization. Expert Syst Appl 33:49–60
Sun XD, Huang RB (2006) Prediction of protein structural classes using support vector machines. Amino Acids 30:469–475
Tan F, Feng X, Fang Z, Li M, Guo Y, Jiang L (2007) Prediction of mitochondrial proteins based on genetic algorithm – partial least squares and support vector machine. Amino Acids 33:669–675
Wang M, Yang J, Chou KC (2005a) Using string kernel to predict signal peptide cleavage site based on subsite coupling model. Amino Acids 28:395–402 (Erratum, ibid. 2005, 29:301)
Wang M, Yang J, Liu GP, Xu ZJ, Chou KC (2004) Weighted-support vector machines for predicting membrane protein types based on pseudo amino acid composition. Protein Eng Ses Sel 17:509–516
Wang M, Yang J, Xu ZJ, Chou KC (2005b) SLLE for predicting membrane protein types. J Theor Biol 232:7–15
Wang SQ, Yang J, Chou KC (2006) Using stacked generalization to predict membrane protein types based on pseudo amino acid composition. J Theor Biol 242:941–946
Wen Z, Li M, Li Y, Guo Y, Wang K (2006) Delaunay triangulation with partial least squares projection to latent structures: a model for G-protein coupled receptors classification and fast structure recognition. Amino Acids 32:277–283
Xiao X, Chou KC (2007) Digital coding of amino acids based on hydrophobic index. Protein Peptide Lett 14:871–875
Xiao X, Shao S, Ding Y, Huang Z, Chen X, Chou KC (2005a) Using cellular automata to generate Image representation for biological sequences. Amino Acids 28:29–35
Xiao X, Shao S, Ding Y, Huang Z, Huang Y, Chou KC (2005b) Using complexity measure factor to predict protein subcellular location. Amino Acids 28:57–61
Xiao X, Shao SH, Ding YS, Huang ZD, Chou KC (2006a) Using cellular automata images and pseudo amino acid composition to predict protein subcellular location. Amino Acids 30:49–54
Xiao X, Shao SH, Huang ZD, Chou KC (2006b) Using pseudo amino acid composition to predict protein structural classes: approached with complexity measure factor. J Comput Chem 27:478–482
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64:643–651
Yuan SF, Chu FL (2007) Fault diagnosis based on support vector machines with parameter optimization by artificial immunization algorithm. Mech Syst Signal Pr 21:1318–1330
Zhang CT, Chou KC (1992) An optimization approach to predicting protein structural class form amino-acid-composition. Protein Sci 1:401–408
Zhang CT, Chou KC, Maggiora GM (1995) Predicting protein structural classes from amino-acid-composition—application of fuzzy Clustering. Protein Eng 8:425–435
Zhang SW, Pan Q, Zhang HC, Shao ZC, Shi JY (2006) Prediction protein homo-oligomer types by pseudo amino acid composition: approached with an improved feature extraction and naive Bayes feature fusion. Amino Acids 30:461–468
Zhang SW, Zhang YL, Yang HF, Zhao CH, Pan Q (2007) Using the concept of Chou’s pseudo amino acid composition to predict protein subcellular localization: an approach by incorporating evolutionary information and von Neumann entropies. Amino Acids. doi:10.1007/s00726-007-0010-9
Zhang TL, Ding YS, Chou KC (2006) Prediction of protein subcellular location using hydrophobic patterns of amino acid sequence. Comput Biolo Chem 30:367–371
Zhang TL, Ding YS (2007) Using pseudo amino acid composition and binary-tree support vector machines to predict protein structural classes. Amino Acids 33:623–629
Zhang TL, Ding YS, Chou KC (2008) Prediction protein structural classes with pseudo-amino acid composition: approximate entropy and hydrophobicity pattern. J Theor Biol 250:186–193
Zhang ZH, Wang ZH, Zhang ZR, Wang YX (2006) A novel method for apoptosis protein subcellular localization prediction combining encoding based on grouped weight and support vector machine. FEBS Lett 580:6169–6174
Zhou GF, Xu XH, Zhang CT (1992) A weighting method for predicting protein structural class form amino-acid-composition. Eur J Biochem 210:747–749
Zhou GP (1998) An intriguing controversy over protein structural class prediction. J Protein Chem 17:729–738
Zhou GP, Assa-Munt N (2001) Some insights into protein structural class prediction. Proteins 44:57–59
Zhou GP, Doctor K (2003) Subcellular location prediction of apoptosis proteins. Proteins 50:44–48
Zhou XB, Chen C, Li ZC, Zou XY (2007) Using Chou’s amphiphilic pseudo-amino acid composition and support vector machine for prediction of enzyme subfamily classes. J Theor Biol 248:546–551
Acknowledgments
The authors acknowledge financial support from the National Natural Science Foundation of China (Nos. 20475068, 20575082), the Natural Science Foundation of Guangdong Province (No. 031577, 7003714), the Scientific Technology Project of Guangdong Province (No. 2005B30101003) and the Scientific Technology Project of Guangzhou City (No. 2007Z3-E0441).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, ZC., Zhou, XB., Lin, YR. et al. Prediction of protein structure class by coupling improved genetic algorithm and support vector machine. Amino Acids 35, 581–590 (2008). https://doi.org/10.1007/s00726-008-0084-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-008-0084-z