Abstract
Introduction
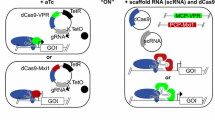
Controlling gene expression is a fundamental goal of basic and synthetic biology because it allows insight into cellular function and control of cellular activity. We explored the possibility of generating an optogenetic repressor of gene expression in the model organism Saccharomyces cerevisiae by using light to control the nuclear localization of nuclease-dead Cas9, dCas9.
Methods
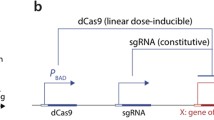
The dCas9 protein acts as a repressor for a gene of interest when localized to the nucleus in the presence of an appropriate guide RNA (sgRNA). We engineered dCas9, the mammalian transcriptional repressor Mxi1, and an optogenetic tool to control nuclear localization (LINuS) as parts in an existing yeast optogenetic toolkit. This allowed expression cassettes containing novel dCas9 repressor configurations and guide RNAs to be rapidly constructed and integrated into yeast.
Results
Our library of repressors displays a range of basal repression without the need for inducers or promoter modification. Populations of cells containing these repressors can be combined to generate a heterogeneous population of yeast with a 100-fold expression range. We find that repression can be dialed modestly in a light dose- and intensity-dependent manner. We used this library to repress expression of the lanosterol 14-alpha-demethylase Erg11, generating yeast with a range of sensitivity to the important antifungal drug fluconazole.
Conclusions
This toolkit will be useful for spatiotemporal perturbation of gene expression in Saccharomyces cerevisiae. Additionally, we believe that the simplicity of our scheme will allow these repressors to be easily modified to control gene expression in medically relevant fungi, such as pathogenic yeasts.











Similar content being viewed by others
References
Anadirekkun, J., C. Stewart, S. Geller, M. Patel, J. Melendez, B. Oakes, M. Noyes, and M. McClean. A yeast optogenetic toolkit (yOTK) for gene expression control in Saccharomyces cerevisiae. BioRxiv 2019. https://doi.org/10.1101/663393.
Aubrey, B., G. Kelly, K. A. M. Brennan, L. O’Connor, L. Milla, S. Wilcox, L. Tai, A. Strasser, and M. Herold. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Reports 10:1422–1432, 2015.
Bódi, Z., Z. Farkas, D. Nevozhay, D. Kalapis, V. Lázár, B. Csörgö, Á. Nyerges, B. Szamecz, G. Fekete, B. Papp, H. Araújo, J. Oliveira, G. Moura, M. Santos, T. Székely, Jr., G. Balázsi, and C. Pál. Phenotypic heterogeneity promotes adaptive evolution. PLOS Biology 15(5):e2000644, 2017.
Brachmann, C., A. Davies, G. Cost, E. Caputo, J. Li, P. Heiter, and J. Boeke. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115–132, 1998.
Branch, M., T. Coleman, and Y. Li. A Subspace, Interior, and Conjugate Gradient Method for Large-Scale Bound-Constrained Minimization Problems. SIAM Journal of Scientific Computing 21(1):1–23, 1999.
Burke, D., D. C. Amberg, and T. Stearns. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Woodbury, NY: Cold Spring Harbor Laboratory Press, 2000.
Davis, K., V. Pattanayak, D. Thompson, J. Zuris, and D. Liu. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nature Chemical Biology 11:316–318, 2015.
Deaner, M., J. Mejia, and H. S. Alper. Enabling graded and large-scale multiplex of desired genes using a dual-mode dCas9 activator in Saccharomyces cerevisiae. ACS Synthetic Biology 6:1931–1943, 2017.
Deltcheva, E., K. Chylinski, C. M. Sharma, K. Gonzales, Y. Chao, Z. A. Pirzada, M. R. Eckert, J. Vogel, and E. Charpentier. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471(7340):602–607, 2011.
Dexter, J., P. Xu, J. Gunawardena, and M. McClean. Robust network structure of the Sln1-Ypd1-Ssk1 three-component phospho-relay prevents unintended activation of the HOG MAPK pathway in Saccharomyces cerevisiae. BMC Systems Biology 9(1):17, 2015.
Dow, L., J. Fisher, K. O’Rourke, A. Muley, E. Kastenhuber, G. Livshits, D. Tschaharganeh, N. Socci, and S. Lowe. Inducible in vivo genome editing with CRISPR-Cas9. Nature Biotechnology 33:390–394, 2015.
Farzadfard, F., S. D. Perli, and T. K. Lu. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synthetic Biology 2(10):604–613, 2013.
Fegan, A., B. White, J. Carlson, and C. Wagner. Chemically controlled protein assembly: Techniques and applications. Chemical Reviews 110:3315–3336, 2010.
Gander, M., J. Vrana, W. Voje, J. Carothers, and E. Klavins. Digital logic circuits in yeast with CRISPR-dCas9 NOR gates. Nature Communications 8:15459, 2017.
Gangopadhyay, S., K. Cox, D. Manna, D. Lim, B. Maji, Q. Zhou, and A. Coudhary. Precision Control of CRISPR-Cas9 Using Small Molecules and Light. Biochemistry 58:234–244, 2019.
Gerhardt, K. P., E. J. Olson, S. M. Castillo-Hair, L. A. Hartsough, B. P. Landry, F. Ekness, R. Yokoo, E. J. Gomez, P. Ramakrishnan, and J. Suh. An open-hardware platform for optogenetics and photobiology. Scientific Reports 6:35363, 2016.
Gietz, R. D., and R. H. Schiestl. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature Protocols 2(1):31–34, 2007.
Gilbert, L. A., M. H. Larson, L. Morsut, Z. Liu, G. A. Brar, S. E. Torres, N. Stern-Ginossar, O. Brandman, E. H. Whitehead, J. A. Doudna, W. A. Lim, and J. S. Weissman. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154(2):442–451, 2013.
Gonzalez, F., Z. Zhu, Z. Shi, K. Lelli, N. Verma, Q. Li, and D. Huangfu. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell 15:215–226, 2014.
Hemphill, J., E. Borchardt, K. Brown, A. Asokan, and A. Deiters. Optical control of CRISPR/Cas9 gene editing. Journal of the American Chemical Society 137:5642–5645, 2015.
Hersen, P., M. McClean, L. Mahadevan, and S. Ramanathan. Signal processing by the HOG MAP kinase pathway. Proceedings of the National Academy of Sciences 105(20):7165–7170, 2008.
Hickman, M. J., and F. Winston. Heme levels switch the function of Hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Molecular and Cellular Biology 27(21):7414–7424, 2007.
Hsu, P., E. Lander, and F. Zhang. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278, 2014.
Jensen, M. Design principles for nuclease-deficient CRISPR-based transcriptional regulators. FEMS Yeast Research 2018. https://doi.org/10.1093/femsyr/foy039.
Jinek, M., K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna, and E. Charpentier. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821, 2012.
E. Jones, E. Oliphant, and P. Peterson. SciPy: Open Source Scientific Tools for Python. 2001. http://www.scipy.org/. Accessed 26 Feb 2019.
Karst, F., and F. Lacrout. Ertosterol biosynthesis in Saccharomyces cerevisiae: mutants deficient in the early steps of the pathway. Molecular Genetics and Genomics 154(3):269–277, 1977.
Kiani, S., J. Beal, M. Ebrahimkhani, J. Huh, R. Hall, Z. Xie, Y. Li, and R. Weiss. CRISPR transcriptional represion devices and layered circuits in mammalian cells. Nature Methods 11:723–726, 2014.
Kleinjan, D., C. Wardrope, S. Nga Sou, and S. Rosser. Drug-tunable multidimensional synthetic gene control using inducible degron-tagged dCas9 effectors. Nature Communications 8:1191, 2017.
Kontoyiannis, D., N. Sagar, and K. Hirschi. Overexpression of Erg11p by the regulatable GAL1 promoter confers fluconazole resistance in Saccharomyces cerevisiae. Antimicrobial Agents and Chemotherapy 43(11):2798–2800, 1999.
Kundert, K., J. Lucas, K. Watters, C. Fellmann, A. Nh, B. Heineike, C. Fitzsimmons, B. Oakes, J. Qu, N. Prasad, O. Rosenberg, D. Savage, H. El-Samad, J. Doudna, and T. Kortemme. Controlling CRISPR-Cas9 with ligand-activated and ligand-deactivated sgRNAs. Nature Communications 10:2127, 2019.
Lawhorn, I., J. Ferreira, and C. Wang. Evaluation of sgRNA target sites for CRISPR-mediated repression of TP53. PloS ONE 9(11):e113232, 2014.
Lee, M. E., W. Deloache, D. Cervantes, and J. Dueber. A highly characterized yeast toolkit for modular, multipart assembly. ACS Synthetic Biology 4(9):975–986, 2015.
Li, S., D. Giardina, and M. Siegal. Control of nongenetic heterogeneity in growth rate and stress tolerance of Saccharomyces cerevisiae by cyclic AMP-regulated transcription factors. PLoS Genetics 14(11):e1007744, 2018.
Lian, J., M. HamediRad, S. Hu, and H. Zhao. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system. Nature Communications 8:1688, 2017.
Liu, K., M. Ramli, C. Woo, Y. Wang, T. Zhao, X. Zhang, G. Yim, B. Chong, A. Gowher, M. Chua, J. Jung, J. Lee, and M. Tan. A chemical-inducible CRISPR–Cas9 system for rapid control of genome editing. Nature Chemical Biology 12:980–987, 2016.
Liu, Y., Y. Zhan, Z. Chen, A. He, J. Li, H. Wu, L. Liu, C. Zhuang, J. Lin, X. Guo, Q. Zhang, W. Huang, and Z. Cai. Directing cellular information flow via CRISPR signal conductors. Nature Methods 13:938–944, 2016.
Ma, H., S. Kunes, P. Schatz, and D. Botstein. Plasmid construction by homologous recombination in yeast. Gene 58:201–216, 1987.
Ma, H., L.-C. Tu, A. Naseri, M. Huisman, S. Zhang, D. Grunwald, and T. Pederson. CRISPR-Cas9 nuclear dynamics and target recognition in living cells. Journal of Cell Biology 214(5):525–537, 2016.
Melendez, J., M. Patel, B. L. Oakes, P. Xu, P. Morton, and M. N. McClean. Real-time optogenetic control of intracellular protein concentration in microbial cell cultures. Integrative Biology 6(3):366–372, 2014.
Natsume, T., and M. Kanemaki. Conditional degrons for controlling protein expression at the protein level. Annual Reviews of Genetics 51:83–102, 2017.
Nevozhay, D., T. Zal, and G. Balazsi. Transferring a synthetic gene circuit from yeast to mammalian cells. Nature Communications 4:1451, 2013.
Nihongaki, Y., F. Kawano, T. Nakajima, and M. Sato. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nature Biotechnology 33:755–760, 2015.
Nihongaki, Y., S. Yamamoto, F. Kawano, H. Suzuki, and M. Sato. CRISPR-Cas9 based photoactivatable transcription system. Chemical Biology 22:169–174, 2015.
Niopek, D., D. Benzinger, J. Roensch, T. Draebing, P. Wehler, R. Eils, and B. DiVentura. Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells. Nature Communications 5:4404, 2014.
Oakes, B., D. Nadler, A. Flamholz, F. Christof, B. Staahl, J. Doudna, and D. Savage. Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nature Biotechnology 34:646–651, 2016.
Oldenburg, K., K. Vo, S. Michaelis, and C. Paddon. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Research 25:451–452, 1997.
Pelet, S., F. Rudolf, M. Nadal-Ribelles, E. de Nadal, F. Posas, and M. Peter. Transient activation of the HOG MAPK pathway regulates bimodal gene expression. Science 332(6030):732–735, 2011.
Polstein, L., and C. Gersbach. A light-inducible CRISPR-Cas9 system for controlling endogenous gene activation. Nature Chemical Biology 11:198–200, 2015.
Qi, L. S., M. H. Larson, L. A. Gilbert, J. A. Doudna, J. S. Weissman, A. P. Arkin, and W. A. Lim. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152(5):1173–1183, 2013.
Rose, J. C., P. S. Huang, N. D. Camp, J. Ye, and A. M. Leidal. A computationally engineered RAS rheostat reveals RAS-ERK signaling dynamics. Nature Chemical Biology 13:119–126, 2017.
Rose, J., J. Stephany, W. Valente, B. Trevillian, H. Dang, J. Bielas, D. Maly, and D. Fowler. Rapidly inducible Cas9 and DSB-ddPCR to probe editing kinetics. Nature Methods 14:891–896, 2017.
Schirmaier, F., and P. Philippsen. Identification of two genes coding for the translation elongation factor EF-1 alpha of S. cerevisiae. EMBO Journal 3(13):3311–3315, 1984.
Schreiber-Agus, N., L. Chin, K. Chen, R. Torres, G. Rao, P. Guida, A. I. Skoultchi, and R. A. DePinho. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell 80(5):777–786, 1995.
Senturk, S., N. Shirole, D. Nowak, V. Corbo, D. Pal, A. Vaughan, D. Tuveson, L. Trotman, J. Kinney, and R. Sordella. Rapid and tunable method to temporally control gene editing based on conditional Cas9 stabilization. Nature Communications 8:14370, 2017.
Sheff, M., and K. Thorn. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21:661–670, 2004.
Siegel, A., M. Baird, M. Davidson, and R. Day. Strengths and Weaknesses of Recently Engineered Red Fluorescent Proteins Evaluated in Live Cells Using Fluorescence Correlation Spectroscopy. International Journal of Molecular Sciences 14:20340–20358, 2013.
Smith, J. D., S. Suresh, U. Schlecht, M. Wu, O. Wagih, G. Peltz, R. W. Davis, L. M. Steinmetz, L. Parts, and R. P. S. Onge. Quantitative CRISPR interference screens in yeast identify chemical-genetic interactions and new rules for guide RNA design. Genome Biology 17:45, 2016.
Strickland, D., X. Yao, G. Gawlak, M. K. Rosen, K. H. Gardner, and T. R. Sosnick. Rationally improving LOV domain–based photoswitches. Nature Methods 7(8):623–626, 2010.
Sweeney, K., N. Moreno Morales, Z. Burmeister, A. Nimunkar, and M. McClean. Easy calibration of the light plate apparatus for optogenetic experiments. MethodsX 6:1480–1488, 2019.
Tang, W., J. Hu, and D. Liu. Aptazyme-embedded guide rnas enable ligand-responsive genome editing and transcriptional activation. Nature Communications 8:15939, 2017.
Vanegas, K., B. Lehka, and U. Mortensen. SWITCH: a dynamic CRISPR tool for genome engineering and metabolic pathway control for cell factory construction in Saccharomyces cerevisiae. Microbial Cell Factories 2017. https://doi.org/10.1186/s12934-017-0632-x.
Winston, F., C. Dollard, and R. Ricupero-Havasse. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53–55, 1995.
Zetsche, B., S. Volz, and F. Zhang. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nature Biotechnology 33:139–142, 2015.
Zwietering, M., I. Jongenburger, F. Rombouts, and K. Van’t Riet. Modeling the bacterial growth curve. Applied and Environmental Microbiology 56(6):1875–1881, 1990.
Acknowledgments
The authors would like to acknowledge discussion and helpful comments from the members of the McClean lab throughout the project. We acknowledge Taylor Scott for help analyzing the growth curve data, Kieran Sweeney for supplying MATLAB code to analyze nuclear localization, and Jidapas (My) An-Adirekkun for assistance with figures. This work was supported by the American Cancer Society [IRG-15-213-51] to M.N.M and by the BMBF 031L0079 grant to B.D.V. Flow cytometry was enabled by the University of Wisconsin Carbone Cancer Center Support Grant P30 CA014520. Megan Nicole McClean, Ph.D., holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Conflict of interest
Authors Stephanie H. Geller, Enoch B. Antwi, Barbara Di Ventura and Megan N. McClean declare that they have no conflicts of interest.
Ethical Standards
No human or animal studies were carried out by the authors for this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Megan N. McClean is an Assistant Professor in the Department of Biomedical Engineering at the University of Wisconsin-Madison. She received her B.A. from the University of California-Berkeley and her Ph.D. from Harvard University, both in Applied Mathematics. During her thesis work with Dr. Sharad Ramanathan, she used computational modeling in combination with single-cell microscopy to understand the mechanisms of crosstalk prevention and signaling specificity in Saccharomyces cerevisiae MAP kinase pathways. Prior to joining UW-Madison, Dr. McClean was a Lewis-Sigler Fellow at Princeton University where she utilized optogenetics, control theory, and synthetic biology to develop tools for controlling biological circuits. At UW-Madison, Dr. McClean’s research group employs systems and synthetic biology approaches to understand biological signal processing in fungi, including human fungal pathogens, with implications for improving treatment strategies. Dr. McClean holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund and a Maximizing Investigators’ Research Award from the National Institute of General Medical Sciences.
This article is part of the CMBE 2019 Young Innovators special issue.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Geller, S.H., Antwi, E.B., Di Ventura, B. et al. Optogenetic Repressors of Gene Expression in Yeasts Using Light-Controlled Nuclear Localization. Cel. Mol. Bioeng. 12, 511–528 (2019). https://doi.org/10.1007/s12195-019-00598-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12195-019-00598-9