Abstract
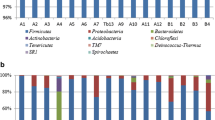
Cheese is one of the traditional fermented dairy products in Xinjiang, China. Due to its geographical location and regional feature this type of cheese harbors certain regional characteristics. To investigate these, here Illumina MiSeq high-throughput sequencing technology was used to target the v4-v5 interval to analyze the composition of fungal flora in Xinjiang traditional cheese. Our results showed the fungal flora of this cheese is mainly composed of Pichia (65.20%), Kazachstania (9.05%), Galactomyces (7.21%), Zygosaccharomyces (6.56%), Torulaspora (3.13%), Dipodascus (2.11%) and Ogataea (1.64%) belonging to the Ascomycota. PcoA (Principal Co-ordinates Analysis) and an UPGMA (unweighted pair-group method with arithmetic means) based on the OTUs (Optical Transform Unit) horizontal-weighted UniFrac distances, revealed some differences in fungal community structure among 17 cheese samples. At the OTU level, nine dominant OTUs were found in all the samples, for which Pichia was the most important fungal group. Building on this, the moisture content (23.20–59.22%), water distribution, and salt content (1.13–4.84%) in cheese were also determined. We found that six of the seven dominant fungal genera had specific correlations with the above physical and chemical variables, with only Ogataea uncorrelated with any variables. The results provide a theoretical basis for the development and use of cheese microbial resources in Xinjiang, China.


Similar content being viewed by others
References
Yang Y, Shevchenko A, Knaust A et al (2014) Proteomics evidence for kefir dairy in Early Bronze Age China. J Archaeol Sci 45:178–186. https://doi.org/10.1016/j.jas.2014.02.005
Zheng X, Fei L, Li K et al (2018) Evaluating the microbial ecology and metabolite profile in Kazak artisanal cheeses from Xinjiang, China. Food Res Int 111:130–136. https://doi.org/10.1016/j.foodres.2018.05.019
Schröder J, Maus I, Trost E, Tauch A (2011) isolated from the surface of smear-ripened cheeses and insights into cheese ripening and flavor generation. BMC Genom 12:545. https://doi.org/10.1186/1471-2164-12-545
Schornsteiner E, Mann E, Bereuter O et al (2014) Cultivation-independent analysis of microbial communities on Austrian raw milk hard cheese rinds. Int J Food Microbiol 180:88–97. https://doi.org/10.1016/j.ijfoodmicro.2014.04.010
Margolies BJ, Barbano DM (2018) Determination of fat, protein, moisture, and salt content of Cheddar cheese using mid-infrared transmittance spectroscopy. J Dairy SCI 101:924–933. https://doi.org/10.3168/jds.2017-13431
Kafili T, Razavi SH, Djomeh ZE et al (2009) Microbial characterization of Iranian traditional Lighvan cheese over manufacturing and ripening via culturing and PCR-DGGE analysis: identification and typing of dominant lactobacilli. Eur Food Res Technol 229:83–92. https://doi.org/10.1007/s00217-009-1028-x
Úbeda JF, Fernández-González M, Briones AI (2009) Application of PCR-TTGE and PCR-RFLP for intraspecific and interspecific characterization of the genus saccharomyces using actin gene (ACT1) Primers. Curr Microbiol 58:58–63. https://doi.org/10.1007/s00284-008-9283-9
Banjara N, Suhr MJ, Hallen-Adams HE (2015) Diversity of yeast and mold species from a variety of cheese types. Curr Microbiol 70:792–800. https://doi.org/10.1007/s00284-015-0790-1
Jingkai J, Jianming Z, Zhenmin L, Huaxi Y (2020) Dynamic changes of microbiota and texture properties during the ripening of traditionally prepared cheese of China. Arch Microbiol 202:2059–2069. https://doi.org/10.1007/10.1007/s00203-020-01921-z
Alejandro A-T, Meyli C et al (2014) High-throughput sequencing of microbial communities in Poro cheese, an artisanal Mexican cheese. Food Microbiol 44:136–141. https://doi.org/10.1016/j.fm.2014.05.022
Chen Y, MacNaughtan W, Jones P et al (2020) The state of water and fat during the maturation of Cheddar cheese. Food Chem 303:1–8. https://doi.org/10.1016/j.foodchem.2019.125390
Beijing: Standards Press of China (2016) GB 5009.44 national standard for food safety determination of chlorine in food. 1–13
Su Z, Cai S, Liu J et al (2021) Root-associated endophytic bacterial community composition of asparagus officinalis of three different varieties. Indian J Microbiol 61:160–169. https://doi.org/10.1007/s12088-021-00926-6
Kuo J, Liu D, Wang SH et al (2021) Dynamic changes in soil microbial communities with glucose enrichment in sediment microbial fuel cells. Indian J Microbiol. https://doi.org/10.1007/s12088-021-00959-x
Rynne NM, Beresford TP, Kelly AL et al (2004) Effect of milk pasteurization temperature and in situ whey protein denaturation on the composition, texture and heat-induced functionality of half-fat Cheddar cheese. Int Dairy J 14:989–1001. https://doi.org/10.1016/j.idairyj.2004.03.010
Gianferri R, D’Aiuto V, Curini R et al (2007) Proton NMR transverse relaxation measurements to study water dynamic states and age-related changes in Mozzarella di Bufala Campana cheese. Food Chem 105:720–726. https://doi.org/10.1016/j.foodchem.2007.01.005
Song L, Shao Y, Ning S et al (2017) Performance of a newly isolated salt-tolerant yeast strain Pichia occidentalis G1 for degrading and detoxifying azo dyes. Bioresour Technol 233:21–29. https://doi.org/10.1016/j.biortech.2017.02.065
Borelli BM, Ferreira EG, Lacerda ICA et al (2006) Yeast populations associated with the artisanal cheese produced in the region of Serra da Canastra, Brazil. World J Microb Biot 22:1115–1119. https://doi.org/10.1007/s11274-006-9151-3
Alegría Á, González R, Díaz M, Mayo B (2011) Assessment of microbial populations dynamics in a blue cheese by culturing and denaturing gradient gel electrophoresis. Curr Microbiol 62:888–893. https://doi.org/10.1007/s00284-010-9799-7
Atanassova MR, Fernandez-Otero C, Rodriguez-Alonso P et al (2016) Characterization of yeasts isolated from artisanal short-ripened cows’ cheeses produced in Galicia (NW Spain). Food Microbiol 53:172–181. https://doi.org/10.1016/j.fm.2015.09.012
Belen C, Johan R et al (2018) Bakery yeasts, a new model for studies in ecology and evolution. Yeast 35:591–603. https://doi.org/10.1002/yea.3350
Nissen S, Faidley TD, Zimmerman DR et al (1994) Colostral milk fat percentage and pig performance are enhanced by feeding the leucine metabolite β-hydroxy β-methylbutyrate to sows. J Anim Sci 72(9):2331–2337. https://doi.org/10.2527/1994.7292331x
Psomas EI, Andrighetto C, Litopoulou-Tzanetaki E et al (2001) Some probiotic properties of yeast isolates from infant faeces and Feta cheese. Int J Food Microbiol 69:125–133. https://doi.org/10.1016/S0168-1605(01)00580-3
Šajbidor J, Lamačka M, Breirerová E et al (1994) Effect of salt stress on fatty acid alterations in some strains of Dipodascopsis and Dipodascus spp. World J Microb Biot 10:184–186. https://doi.org/10.1007/BF00360883
Kuroda K, Kitagawa Y, Kobayashi K et al (2006) Antibody production by a protease-deficient strain of methylotrophic yeast. Ogataea minuta Microb Cell Factories 5:64. https://doi.org/10.1186/1475-2859-5-S1-P64
Acknowledgements
We would like to thank the Northwest Hubei Research Institute of Traditional Fermented Food for their technical support. This research was supported by the Natural Science Foundation of China [grant number 32060548]; the Key Scientific and Technological Research Projects in the Key Areas of the Xinjiang Production and Construction Corps [grant numbers 2018AB010, 2018AB015]; and the Open Project Program of Key Laboratory of Xinjiang Phytomedicine Resource and Utilization of Ministry of Education [grant number XPRU202007].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they do not have any conflict of interest.
Human and Animal Rights
This study does not involve any human or animal testing.
Informed Consent
Written informed consent was obtained from all study participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, K., Zhang, Y., Li, S. et al. Fungal Diversity in Xinjiang Traditional Cheese and its Correlation With Moisture Content. Indian J Microbiol 62, 47–53 (2022). https://doi.org/10.1007/s12088-021-00967-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-021-00967-x