Abstract
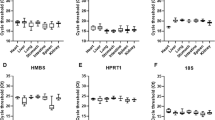
The precise estimation of postmortem interval (PMI) is a critical step in death investigation of forensic cases. Detecting the degradation of RNA in tissues by real time quantitative polymerase chain reaction (RT-qPCR) technology provides a new theoretical basis for estimation of PMI. However, most commonly used reference genes degrade over time, while previous studies seldom consider this when selecting suitable reference genes for the estimation of PMI. Studies have shown microRNAs (miRNAs) are very stable and circular RNAs (circRNAs) have recently emerged as a novel class of RNAs with high stability. We aimed to evaluate the stability of the two kinds of RNAs and normal reference genes using geNorm and NormFinder algorithms to identify tissue-specific reference genes for PMI estimation. The content of candidate RNAs from mouse heart, liver and skeletal muscle tissues were dynamically examined in 8 consecutive days after death. Among the 11 candidate genes (β-actin, Gapdh, Rps18, 5S, 18S, U6, miR-133a, miR-122, circ-AFF1, LC-Ogdh and LC-LRP6), the following genes showed prioritized stability: miR-122, miR-133a and 18S in heart tissues; LC-Ogdh, circ-AFF1 and miR-122 in liver tissues; and miR-133a, circ-AFF1 and LC-LRP6 in skeletal muscle tissues. Our results suggested that miRNAs and circRNAs were more stable as reference genes than other kinds of RNAs regarding PMI estimation. The appropriate internal control genes were not completely the same across tissue types.



Similar content being viewed by others
References
Lee DG, Yang KE, Hwang JW, Kang HS, Lee SY, Choi S, et al. Degradation of kidney and psoas muscle proteins as indicators of post-mortem interval in a rat model, with use of lateral flow technology. PLoS One. 2016;11:e160557.
van den Berge M, Wiskerke D, Gerretsen RR, Tabak J, Sijen T. DNA and RNA profiling of excavated human remains with varying postmortem intervals. Int J Legal Med. 2016;130:1471–80.
Lv YH, Ma JL, Pan H, Zeng Y, Tao L, Zhang H, et al. Estimation of the human postmortem interval using an established rat mathematical model and multi-RNA markers. Forensic Sci Med Pathol. 2017;13:20–7.
Arya M, Shergill IS, Williamson M, Gommersall L, Arya N, Patel HR. Basic principles of real-time quantitative PCR. Expert Rev Mol Diagn. 2005;5:209–19.
Young ST, Wells JD, Hobbs GR, Bishop CP. Estimating postmortem interval using RNA degradation and morphological changes in tooth pulp. Forensic Sci Int. 2013;229:161–3.
Ma J, Pan H, Zeng Y, Lv Y, Zhang H, Xue A, et al. Exploration of the R code-based mathematical model for PMI estimation using profiling of RNA degradation in rat brain tissue at different temperatures. Forensic Sci Med Pathol. 2015;11:530–7.
Birdsill AC, Walker DG, Lue L, Sue LI, Beach TG. Postmortem interval effect on RNA and gene expression in human brain tissue. Cell Tissue Bank. 2011;12:311–8.
Lv YH, Ma JL, Pan H, Zhang H, Li WC, Xue AM, et al. RNA degradation as described by a mathematical model for postmortem interval determination. J Forensic Legal Med. 2016;44:43–52.
Gonzalez-Herrera L, Valenzuela A, Marchal JA, Lorente JA, Villanueva E. Studies on RNA integrity and gene expression in human myocardial tissue, pericardial fluid and blood, and its postmortem stability. Forensic Sci Int. 2013;232:218–28.
Poor VS, Lukacs D, Nagy T, Racz E, Sipos K. The rate of RNA degradation in human dental pulp reveals post-mortem interval. Int J Legal Med. 2016;130:615–9.
Li W, Ma K, Lv Y, Zhang P, Pan H, Zhang H, et al. Postmortem interval determination using 18S-rRNA and microRNA. Sci Justice. 2014;54:307–10.
Heinrich M, Lutz-Bonengel S, Matt K, Schmidt U. Real-time PCR detection of five different “endogenous control gene” transcripts in forensic autopsy material. Forensic Sci Int Genet. 2007;1:163–9.
Jung HJ, Suh Y. Circulating miRNAs in ageing and ageing-related diseases. J Genet Genomics. 2014;41:465–72.
Lv Y, Ma K, Zhang H, He M, Zhang P, Shen Y, et al. A time course study demonstrating mRNA, microRNA, 18S rRNA, and U6 snRNA changes to estimate PMI in deceased rat's spleen. J Forensic Sci. 2014;59:1286–94.
Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta (BBA) - Gene Regulatory Mech. 2016;1859:163–8.
Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer Tokyo. 2017. 2018;25:1–7.
Zhu M, Xu Y, Chen Y, Yan F. Circular BANP, an upregulated circular RNA that modulates cell proliferation in colorectal cancer. Biomed Pharmacother. 2017;88:138–44.
Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular carcinoma development. Medicine (Baltimore). 2016;95:e3811.
Zhao Z, Li X, Jian D, Hao P, Rao L, Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54:237–45.
Werfel S, Nothjunge S, Schwarzmayr T, Strom T, Meitinger T, Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol. 2016;98:103–7.
Zhang Y, Liu B, Shao C, Xu H, Xue A, Zhao Z, et al. Evaluation of the inclusion of circular RNAs in mRNA profiling in forensic body fluid identification. Int J Legal Med. 2018;132:43–52.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:H34.
Andersen CL, Jensen JL, Rntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50.
Zhu Y, Wang L, Yin Y, Yang E. Systematic analysis of gene expression patterns associated with postmortem interval in human tissues. Sci Rep. 2017;7:5435.
Ghani M, Sato C, Rogaeva E. Segmental duplications in genome-wide significant loci and housekeeping genes; warning for GAPDH and ACTB. Neurobiol Aging. 2013;34:1710–1.
Gerstberger S, Meyer C, Benjamin-Hong S, Rodriguez J, Briskin D, Bognanni C, et al. The conserved RNA exonuclease Rexo5 is required for 3′ end maturation of 28S rRNA, 5S rRNA, and snoRNAs. Cell Rep. 2017;21:758–72.
Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–52.
Koppelkamm A, Vennemann B, Fracasso T, Lutz-Bonengel S, Schmidt U, Heinrich M. Validation of adequate endogenous reference genes for the normalisation of qPCR gene expression data in human post mortem tissue. Int J Legal Med. 2010;124:371–80.
Ling D, Salvaterra PM. Robust RT-qPCR data normalization: validation and selection of internal reference genes during post-experimental data analysis. PLoS One. 2011;6:e17762.
Wang Q, Ishikawa T, Michiue T, Zhu B, Guan D, Maeda H. Stability of endogenous reference genes in postmortem human brains for normalization of quantitative real-time PCR data: comprehensive evaluation using geNorm, NormFinder, and BestKeeper. Int J Legal Med. 2012;126:943–52.
Meijerink J, Mandigers C, van de Locht L, Tonnissen E, Goodsaid F, Raemaekers J. A novel method to compensate for different amplification efficiencies between patient DNA samples in quantitative real-time PCR. J Mol Diagn. 2001;3:55–61.
Guenin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, Gutierrez L. Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot. 2009;60:487–93.
Neville MJ, Collins JM, Gloyn AL, McCarthy MI, Karpe F. Comprehensive human adipose tissue mRNA and microRNA endogenous control selection for quantitative real-time-PCR normalization. Obesity (Silver Spring). 2011;19:888–92.
Alrowaithi MA, McCallum NA, Watson ND. A method for determining the age of a bloodstain. Forensic Sci Int. 2014;234:e30–1.
Nguyen THD, Galej WP, Bai X, Savva CG, Newman AJ, Scheres SHW, et al. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature. 2015;523:47–52.
Shi C, Yang F, Zhu X, Du E, Yang Y, Wang S, et al. Evaluation of housekeeping genes for quantitative real-time PCR analysis of Bradysia odoriphaga (Diptera: Sciaridae). Int J Mol Sci. 2016;17:1034.
Funding
This work was financially sponsored by the Opening Foundation of Shanghai Key Laboratory of Crime Scene Evidence (grant No.XCWZ20).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflict ofinterest. The authors want to thank Professor Jian-Hui Xie from the Department of Forensic Medicine, School of Basic Medical Sciences, Fudan University for his assistance in screening potential circRNAs.
Ethical approval
All applicable international, national, and/or institutionalguidelines for the care and use of animals were followed. Allprocedures performed in studies involving animals were in accordance with the Ethical Review Board at the School of Basic Medical Sciences, Fudan University.
Rights and permissions
About this article
Cite this article
Tu, C., Du, T., Shao, C. et al. Evaluating the potential of housekeeping genes, rRNAs, snRNAs, microRNAs and circRNAs as reference genes for the estimation of PMI. Forensic Sci Med Pathol 14, 194–201 (2018). https://doi.org/10.1007/s12024-018-9973-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12024-018-9973-y