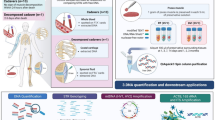
Abstract
When postmortem intervals (PMIs) increase such as with longer burial times, human remains suffer increasingly from the taphonomic effects of decomposition processes such as autolysis and putrefaction. In this study, various DNA analysis techniques and a messenger RNA (mRNA) profiling method were applied to examine for trends in nucleic acid degradation and the postmortem interval. The DNA analysis techniques include highly sensitive DNA quantitation (with and without degradation index), standard and low template STR profiling, insertion and null alleles (INNUL) of retrotransposable elements typing and mitochondrial DNA profiling. The used mRNA profiling system targets genes with tissue specific expression for seven human organs as reported by Lindenbergh et al. (Int J Legal Med 127:891-900, 27) and has been applied to forensic evidentiary traces but not to excavated tissues. The techniques were applied to a total of 81 brain, lung, liver, skeletal muscle, heart, kidney and skin samples obtained from 19 excavated graves with burial times ranging from 4 to 42 years. Results show that brain and heart are the organs in which both DNA and RNA remain remarkably stable, notwithstanding long PMIs. The other organ tissues either show poor overall profiling results or vary for DNA and RNA profiling success, with sometimes DNA and other times RNA profiling being more successful. No straightforward relations were observed between nucleic acid profiling results and the PMI. This study shows that not only DNA but also RNA molecules can be remarkably stable and used for profiling of long-buried human remains, which corroborate forensic applications. The insight that the brain and heart tissues tend to provide the best profiling results may change sampling policies in identification cases of degrading cadavers.
Similar content being viewed by others
References
Goff ML (2009) Early post-mortem changes and stages of decomposition in exposed cadavers. Exp Appl Acarol 49(1–2):21–36
Hau TC, Hamzah NH, Lian HH, Hamzah SPAA (2014) Decomposition process and post mortem changes: review. Sains Malaysiana 43(12):1873–1882
Johnson LA, Ferris JA (2002) Analysis of postmortem DNA degradation by single-cell gel electrophoresis. Forensic Sci Int 126(1):43–47
Calacal GC, Apaga DLT, Salvador JM, Jimenez JAD, Lagat LJ, Villacorta RPF, De Ungria MCA (2015) Comparing different post-mortem human samples as DNA sources for downstream genotyping and identification. Forensic Sci Int Genet 19:212–220
Courts C, Sauer E, Hofmann Y, Madea B, Schyma C (2015) Assessment of STR typing success rate in soft tissues from putrefied bodies based on a quantitative grading system for putrefaction. J Forensic Sci 60(4):1016–1021
Hansen J, Lesnikova I, Funder AMD, Banner J (2014) DNA and RNA analysis of blood and muscle from bodies with variable postmortem intervals. Forensic Sci Med Pathol 10(3):322–328
Hoff-Olsen P, Mevåg B, Staalstrøm E, Hovde B, Egeland T, Olaisen B (1999) Extraction of DNA from decomposed human tissue: an evaluation of five extraction methods for short tandem repeat typing. Forensic Sci Int 105(3):171–183
Ogata M, Mattern R, Schneider PM, Schacker U, Kaufmann T, Rittner C (1990) Quantitative and qualitative analysis of DNA extracted from postmortem muscle tissues. Zeitschrift für Rechtsmedizin 103(6):397–406
Itani M, Yamamoto Y, Doi Y, Miyaishi S (2011) Quantitative analysis of DNA degradation in the dead body. Acta Med Okayama 65(5):299–306
Pooniya S, Lalwani S, Raina A, Millo T, Dogra TD (2014) Quality and quantity of extracted deoxyribonucleic acid (DNA) from preserved soft tissues of putrefied unidentifiable human corpse. J Lab Phys 6(1):31
Schwark T, Heinrich A, von Wurmb-Schwark N (2011) Genetic identification of highly putrefied bodies using DNA from soft tissues. Int J Legal Med 125(6):891–894
Kaiser C, Bachmeier B, Conrad C, Nerlich A, Bratzke H, Eisenmenger W, Peschel O (2008) Molecular study of time dependent changes in DNA stability in soil buried skeletal residues. Forensic Sci Int 177(1):32–36
Ebuehi OA, Amode M, Balogun A, Fowora A (2015) Postmortem time affects brain, liver, kidney and heart DNA in male rat. Am J Biochem 5(1):1–5
Ludes B, Pfitzinger H, Mangin P (1993) DNA fingerprinting from tissues after variable postmortem periods. J Forensic Sci 38(3):686–690
Bär W, Kratzer A, Mächler M, Schmid W (1988) Postmortem stability of DNA. Forensic Sci Int 39(1):59–70
de Leeuwe R, Groen WJM, A taphonomic study based on observations of 196 exhumations and 23 clandestine burials, Taphonomy of human remains: Forensic analysis of the dead and the depositional environment. E. Schotmans, N. Márquez-Grant, S. Forbes, Wiley-Blackwell, in press.
Koppelkamm A, Vennemann B, Lutz-Bonengel S, Fracasso T, Vennemann M (2011) RNA integrity in post-mortem samples: influencing parameters and implications on RT-qPCR assays. Int J Legal Med 125(4):573–580
Sampaio-Silva F, Magalhães T, Carvalho F, Dinis-Oliveira RJ, Silvestre R (2013) Profiling of RNA degradation for estimation of post morterm interval. PLoS One 8(2):e56507
Sijen T (2015) Molecular approaches for forensic cell type identification: on mRNA, miRNA, DNA methylation and microbial markers. Forensic Sci Int Genet 18:21–32
Lindenbergh A, de Pagter M, Ramdayal G, Visser M, Zubakov D, Kayser M, Sijen T (2012) A multiplex (m) RNA-profiling system for the forensic identification of body fluids and contact traces. Forensic Sci Int Genet 6(5):565–577
van den Berge M et al (2014) A collaborative European exercise on mRNA-based body fluid/skin typing and interpretation of DNA and RNA results. Forensic Sci Int Genet 10:40–48
Haas C, Klesser B, Maake C, Bär W, Kratzer A (2009) mRNA profiling for body fluid identification by reverse transcription endpoint PCR and realtime PCR. Forensic Sci Int Genet 3(2):80–88
Hanson EK, Ballantyne J (2013) Highly specific mRNA biomarkers for the identification of vaginal secretions in sexual assault investigations. Sci Justice 53(1):14–22
Juusola J, Ballantyne J (2005) Multiplex mRNA profiling for the identification of body fluids. Forensic Sci Int 152(1):1–12
Roeder AD, Haas C (2013) mRNA profiling using a minimum of five mRNA markers per body fluid and a novel scoring method for body fluid identification. Int J Legal Med 127(4):707–721
Harteveld J, Lindenbergh A, Sijen T (2013) RNA cell typing and DNA profiling of mixed samples: can cell types and donors be associated? Sci Justice 53(3):261–269
Lindenbergh A, van den Berge M, Oostra RJ, Cleypool C, Bruggink A, Kloosterman A, Sijen T (2013) Development of a mRNA profiling multiplex for the inference of organ tissues. Int J Legal Med 127(5):891–900
Bauer M, Gramlich I, Polzin S, Patzelt D (2003) Quantification of mRNA degradation as possible indicator of postmortem interval—a pilot study. Legal Med 5(4):220–227
Heinrich M, Matt K, Lutz-Bonengel S, Schmidt U (2007) Successful RNA extraction from various human postmortem tissues. Int J Legal Med 121(2):136–142
Koppelkamm A, Vennemann B, Fracasso T, Lutz-Bonengel S, Schmidt U, Heinrich M (2010) Validation of adequate endogenous reference genes for the normalisation of qPCR gene expression data in human post mortem tissue. Int J Legal Med 124(5):371–380
Poór VS, Lukács D, Nagy T, Rácz E, Sipos K (2016) The rate of RNA degradation in human dental pulp reveals post-mortem interval. Int J Legal Med 130(3):615–619
Maeda H, Zhu BL, Ishikawa T, Michiue T (2010) Forensic molecular pathology of violent deaths. Forensic Sci Int 203(1):83–92
Nicklas JA, Buel E (2006) Simultaneous determination of total human and male DNA using a duplex real‐time PCR assay. J Forensic Sci 51(5):1005–1015
Pineda GM, Montgomery AH, Thompson R, Indest B, Carroll M, Sinha SK (2014) Development and validation of innoQuant™, a sensitive human DNA quantitation and degradation assessment method for forensic samples using high copy number mobile elements Alu and SVA. Forensic Sci Int Genet 13:224–235
Sinha S, Murphy G, Brown H, Montgomery A, Carrol M, Tabak J (2015) Retrotransposable elements: novel and sensitive DNA markers and their application in human identity. Forensic Sci Int Genet Suppl Ser 5:e627–e629
LaRue BL, Sinha SK, Montgomery AH, Thompson R, Klaskala L, Ge J, King M, Turnbough M, Budowle B (2012) INNULs: a novel design amplification strategy for retrotransposable elements for studying population variation. Hum Hered 74(1):27–35
Weiler NEC, de Vries G, Sijen T (2016) Development of a control region-based mtDNA SNaPshot™ selection tool, integrated into a mini amplicon sequencing method. Sci Justice 56(2):96–103
Clark MA, Worrell MB, Pless JE (1997) Postmortem changes in soft tissues, Forensic Taphonomy; W.D. Haglund, M.H. Sorg, CRC Press 161
Ewing MM, Thompson JM, McLaren RS, Purpero VM, Thomas KJ, Dobrowski PA, DeGroot GA, Romsos EL, Storts DR (2016) Human DNA quantification and sample quality assessment: developmental validation of the PowerQuant® system. For Forensic Sci Int Genet 23:166–177
Westen AA, Grol LJ, Harteveld J, Matai AS, de Knijff P, Sijen T (2012) Assessment of the stochastic threshold, back-and forward stutter filters and low template techniques for NGM. Forensic Sci Int Genet 6(6):708–715
Eichmann C, Parson W (2008) ‘Mitominis’: multiplex PCR analysis of reduced size amplicons for compound sequence analysis of the entire mtDNA control region in highly degraded samples. Int J Legal Med 122(5):385–388
Berger C, Parson W (2009) Mini-midi-mito: adapting the amplification and sequencing strategy of mtDNA to the degradation state of crime scene samples. Forensic Sci Int Genet 3(3):149–153
Lindenbergh A, Maaskant P, Sijen T (2013) Implementation of RNA profiling in forensic casework. Forensic Sci Int Genet 7(1):159–166
Foran DR (2006) Relative degradation of nuclear and mitochondrial DNA: an experimental approach. J Forensic Sci 51(4):766–770
Lee J, Hever A, Willhite D, Zlotnik A, Hevezi P (2005) Effects of RNA degradation on gene expression analysis of human postmortem tissues. FASEB J 19(10):1356–1358
Mitchell PS, Espy MJ, Smith TF, Toal DR, Rys PN, Berbari EF, Osmon DR, Persing DH (1997) Laboratory diagnosis of central nervous system infections with herpes simplex virus by PCR performed with cerebrospinal fluid specimens. J Clin Microbiol 35(11):2873–2877
Ratnamohan VM, Cunningham AL, Rawlinson WD (1998) Removal of inhibitors of CSF-PCR to improve diagnosis of herpesviral encephalitis. J Virol Methods 72(1):59–65
D’Erchia AM, Atlante A, Gadaleta G, Pavesi G, Chiara M, De Virgilio C, Gissi C (2015) Tissue-specific mtDNA abundance from exome data and its correlation with mitochondrial transcription, mass and respiratory activity. Mitochondrion 20:13–21
Fordyce SL, Kampmann ML, Van Doorn NL, Gilbert MTP (2013) Long-term RNA persistence in postmortem contexts. Investig Genet 4(1):1
Lindahl T (1993) Instability and decay of the primary structure of DNA. Nature 362(6422):709–715
Varani G, McClain WH (2000) The G·U wobble base pair. EMBO Rep 1(1):18–23
Suay L, Salvador ML, Abesha E, Klein U (2005) Specific roles of 5′ RNA secondary structures in stabilizing transcripts in chloroplasts. Nucleic Acids Res 33(15):4754–4761
Mitchell P, Tollervey D (2001) mRNA turnover. Curr Opin Cell Biol 13(3):320–325
Birdsill AC, Walker DG, Lue L, Sue LI, Beach TG (2011) Postmortem interval effect on RNA and gene expression in human brain tissue. Cell Tissue Bank 12(4):311–318
Cummings TJ, Strum JC, Yoon LW, Szymanski MH, Hulette CM (2001) Recovery and expression of messenger RNA from postmortem human brain tissue. Mod Pathol 14(11):1157–1161
Preece P, Cairns NJ (2003) Quantifying mRNA in postmortem human brain: influence of gender, age at death, postmortem interval, brain pH, agonal state and inter-lobe mRNA variance. Mol Brain Res 118(1):60–71
van den Berge M, Bhoelai B, Harteveld J, Matai A, Sijen T (2016) Advancing forensic RNA typing: on non-target secretions, a nasal mucosa marker, a differential co-extraction protocol and the sensitivity of DNA and RNA profiling. Forensic Sci Int Genet 20:119–129
Acknowledgments
The authors are grateful to all the donors from whom tissues have been used in this study. We thank Natalie Weiler (Netherlands Forensic Institute), Gina Pineda (InnoGenomics) and Sudhir Sinha (InnoGenomics) for technical assistance. Frank van de Goot and W.J. Mike Groen are thanked for sample collection and providing information on the exhumed bodies. Corina Benschop is thanked for critically reading the manuscript. TS and MvdB received financial support from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 285487 (EUROFORGEN-NoE).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
van den Berge, M., Wiskerke, D., Gerretsen, R.R.R. et al. DNA and RNA profiling of excavated human remains with varying postmortem intervals. Int J Legal Med 130, 1471–1480 (2016). https://doi.org/10.1007/s00414-016-1438-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-016-1438-9