Abstract
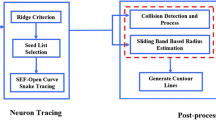
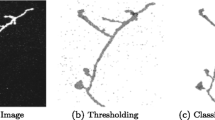
The automatic neuron reconstruction is important since it accelerates the collection of 3D neuron models for the neuronal morphological studies. The majority of the previous neuron reconstruction methods only focused on tracing neuron fibres without considering the somatic surface. Thus, topological errors often present around the soma area in the results obtained by these tracing methods. Segmentation of the soma structures can be embedded in the existing neuron tracing methods to reduce such topological errors. In this paper, we present a novel method to segment the soma structures with complex geometry. It can be applied along with the existing methods in a fully automated pipeline. An approximate bounding block is firstly estimated based on a geodesic distance transform. Then the soma segmentation is obtained by evolving the surface with a set of morphological operators inside the initial bounding region. By evaluating the methods against the challenging images released by the BigNeuron project, we showed that the proposed method can outperform the existing soma segmentation methods regarding the accuracy. We also showed that the soma segmentation can be used for enhancing the results of existing neuron tracing methods.













Similar content being viewed by others
References
Acciai, L, Costantini, I, Pavone, FS, Conti, V, Guerrini, R, Soda, P, & Iannello, G. (2016a). Towards automated neuron tracing via global and local 3D image analysis. In: 2016 IEEE International Symposium on Biomedical Imaging (ISBI), pp. 322– 325.
Acciai, L, Soda, P, & Iannello, G. (2016b). Automatic neuron tracing using a locally tunable approach. In: 2016 IEEE International Symposium on Computer-Based Medical Systems (CBMS), pp. 130–135.
Álvarez, L, Baumela, L, Henríquez, P, & Márquez-Neila, P. (2010). Morphological snakes. In: 2010 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2197–2202.
Basu, S, & Racoceanu, D. (2014). Reconstructing neuronal morphology from microscopy stacks using fast marching. In: 2014 IEEE International Conference on Image Processing (ICIP), pp. 3597–3601.
Basu, S, Ooi, W T, & Racoceanu, D. (2016). Neurite tracing with object process. IEEE Transactions on Medical Imaging, 35(6), 1443–1451.
Chan, T F, & Vese, L A. (2001). Active contours without edges. IEEE Transactions, on Image Processing, 10(2), 266–277.
Chen, H, Xiao, H, Liu, T, & Peng, H. (2015). Smarttracing: self-learning-based neuron reconstruction. Brain Informatics, 2(3), 135–144.
Feng, L, Zhao, T, & Kim, J. (2015). NeuTube 1.0: A new design for efficient neuron reconstruction software based on the swc format. eNeuro, 2(1), 0049.
Gala, R, Chapeton, J, Jitesh, J, Bhavsar, C, & Stepanyants, A. (2014). Active learning of neuron morphology for accurate automated tracing of neurites. Frontiers in Neuroanatomy, 8, 37.
Gulyanon, S, Sharifai, N, Bleykhman, S, Kelly, E, Kim, M, Chiba, A, & Tsechpenakis, G. (2015). Three-dimensional neurite tracing under globally varying contrast. In: 2015 IEEE international symposium on biomedical imaging (ISBI), pp 875–879.
Gulyanon, S, Sharifai, N, Kim, MD, Chiba, A, & Tsechpenakis, G. (2016). CRF Formulation of active contour population for efficient three-dimensional neurite tracing. In: 2016 IEEE international symposium on biomedical imaging (ISBI), pp. 593–597.
Hassouna, M S, & Farag, A A. (2007). Multistencils fast marching methods: a highly accurate solution to the eikonal equation on cartesian domains. IEEE Transactions on Pattern Analysis and Machine Intelligence, 29(9), 1563–1574.
Kayasandik, C B, & Labate, D. (2016). Improved detection of soma location and morphology in fluorescence microscopy images of neurons. Journal of Neuroscience Methods, 274, 61–70.
Labate, D, Laezza, F, Negi, P, Ozcan, B, & Papadakis, M. (2014). Efficient processing of fluorescence images using directional multiscale representations. Mathematical Modelling of Natural Phenomena, 9(5), 177–193.
Liu, S, Zhang, D, Liu, S, Feng, D, Peng, H, & Cai, W. (2016). Rivulet: 3d neuron morphology tracing with iterative back-tracking . Neuroinformatics, 14(4), 387–401.
Liu, S, Zhang, D, Song, Y, Peng, H, & Cai, W. (2017). Automated 3D neuron tracing with precise branch erasing and confidence controlled back-tracking. bioRxiv, p. 109892.
Marquez-Neila, P, Baumela, L, & Alvarez, L. (2014). A morphological approach to curvature-based evolution of curves and surfaces. IEEE Transactions on Pattern Analysis and Machine Intelligence, 36(1), 2–17.
Maturana, M I, Kameneva, T, Burkitt, A N, Meffin, H, & Grayden, D B. (2014). The effect of morphology upon electrophysiological responses of retinal ganglion cells: simulation results. Journal of Computational Neuroscience, 36(2), 157–175.
Ming, X, Li, A, Wu, J, Yan, C, Ding, W, Gong, H, Zeng, S, & Liu, Q. (2013). Rapid reconstruction of 3D neuronal morphology from light microscopy images with augmented rayburst sampling. PLoS One, 8(12), e84,557.
Ozcan, B, Negi, P, Laezza, F, Papadakis, M, & Labate, D. (2015). Automated detection of soma location and morphology in neuronal network cultures. PLoS One, 10(4), e0121,886.
Parekh, R, & Ascoli, G A. (2013). Neuronal morphology goes digital: a research hub for cellular and system neuroscience. Neuron, 77(6), 1017–1038.
Peng, H, Long, F, & Myers, G. (2011). Automatic 3D neuron tracing using all-path pruning. Bioinformatics, 27(13), i239–i247.
Peng, H, Roysam, B, & Ascoli, G A. (2013). Automated image computing reshapes computational neuroscience. BMC Bioinformatics, 14(1), 293.
Peng, H, Meijering, E, & Ascoli, G. (2015). From DIADEM to BigNeuron. Neuroinformatics, 13(3), 259–260.
Pieper, S, Halle, M, & Kikinis, R. (2004). 3D Slicer. In: 2004 IEEE international symposium on biomedical imaging, pp 632–635.
Santamaría-Pang, A, Hernandez-Herrera, P, Papadakis, M, Saggau, P, & Kakadiaris, I A. (2015). Automatic morphological reconstruction of neurons from multiphoton and confocal microscopy images using 3D tubular models. Neuroinformatics, 13(3), 297–320.
Sironi, A, Tekin, B, Rigamonti, R, Lepetit, V, & Fua, P. (2015). Learning separable filters. IEEE Transactions on Pattern Analysis and Machine Intelligence, 37(1), 94–106.
Sironi, A, Türetken, E, Lepetit, V, & Fua, P. (2016). Multiscale centerline detection. IEEE Transactions on Pattern Analysis and Machine Intelligence, 38(7), 1327–1341.
Wang, Y, Narayanaswamy, A, Tsai, C L, & Roysam, B. (2011). A broadly applicable 3-D neuron tracing method based on open-curve snake. Neuroinformatics, 9(2–3), 193–217.
Xiao, H, & Peng, H. (2013). APP2: Automatic tracing of 3D neuron morphology based on hierarchical pruning of a gray-weighted image distance-tree. Bioinformatics, 29(11), 1448–1454.
Yan, C, Li, A, Zhang, B, Ding, W, Luo, Q, & Gong, H. (2013). Automated and accurate detection of soma location and surface morphology in large-scale 3D neuron images. PLoS One, 8(4), e62,579.
Zhang, D, Liu, S, Liu, S, Feng, D, Peng, H, & Cai, W. (2016). Reconstruction of 3D neuron morphology using rivulet back-tracking. In: 2016 IEEE international symposium on biomedical imaging (ISBI), pp 598–601.
Zhao, T, Xie, J, Amat, F, Clack, N, Ahammad, P, Peng, H, Long, F, & Myers, E. (2011). Automated reconstruction of neuronal morphology based on local geometrical and global structural models. Neuroinformatics, 9(2–3), 247–261.
Zhou, Z, Liu, X, Long, B, & Peng, H. (2016). Tremap: Automatic 3D neuron reconstruction based on tracing, reverse mapping and assembling of 2D projections. Neuroinformatics, 14(1), 41– 50.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zhang, D., Liu, S., Song, Y. et al. Automated 3D Soma Segmentation with Morphological Surface Evolution for Neuron Reconstruction. Neuroinform 16, 153–166 (2018). https://doi.org/10.1007/s12021-017-9353-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-017-9353-x