Abstract
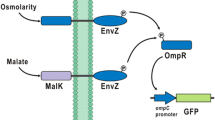
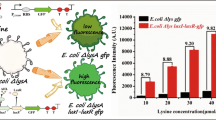
In this study, we constructed amino acid biosensors that can be used as a high-throughput system to screen microorganisms that produce glutamate. The biosensors are based on two-component regulatory systems (TCRSs) combined with green fluorescent protein (GFP) as a reporter. A chimeric DegS/EnvZ (DegSZ) TCRS was constructed by fusing the N-terminal domain of the sensor kinase DegS from Planococcus sp. PAMC21323 with the catalytic domain of the osmosensor EnvZ from Escherichia coli to control expression of gfp in response to glutamate. gfp was controlled by the ompC promoter through the activated response regulator OmpR-P. The chimeric TCRS-based biosensors showed a 4-fold increase in the fluorescent signal after adding glutamate. A linear correlation was observed between fluorescence intensity and exogenously added glutamate concentration. The chimeric TCRS-based biosensor was used to determine glutamate concentration at the single-cell level by fluorescence-activated cell sorting. Therefore, this biosensor can be used to isolate novel gene products and optimize pathways involved in amino acid production.







Similar content being viewed by others
References
Ikeda, M. (2003). In R. Faurie (Ed.), Microbial Production of l-Amino Acids. Advances in Biochemical Engineering/Biotechnology, vol 79: amino acid production process. Berlin: Springer.
Park, J. H., & Lee, S. Y. (2008). Towards systems metabolic engineering of microorganisms for amino acid production. Current Opinion in Biotechnology, 19(5), 454–460. https://doi.org/10.1016/j.copbio.2008.08.007.
Rangan, C., & Barceloux, D. G. (2009). Food additives and sensitivities. a-Month, 55(5), 292–311. https://doi.org/10.1016/j.disamonth.2009.01.004.
Lee, J. Y., Na, Y. A., Kim, E., Lee, H. S., & Kim, P. (2016). The actinobacterium Corynebacterium glutamicum, an industrial workhorse. Journal of Microbiology and Biotechnology, 26(5), 807–822. https://doi.org/10.4014/jmb.1601.01053.
Choi, J. W., Yim, S. S., Lee, S. H., Kang, T. J., Park, S. J., & Jeong, K. J. (2015). Enhanced production of gamma-aminobutyrate (GABA) in recombinant Corynebacterium glutamicum by expressing glutamate decarboxylase active in expanded pH range. Microbial Cell Factories, 14(1), 1–11. https://doi.org/10.1186/s12934-015-0205-9.
Park, S. J., Kim, E. Y., Noh, W., Oh, Y. H., Kim, H. Y., Song, B. K., Cho, K. M., Hong, S. H., Lee, S. H., & Jegal, J. (2013). Synthesis of nylon 4 from gamma-aminobutyrate (GABA) produced by recombinant Escherichia coli. Bioprocess and Biosystems Engineering, 36(7), 885–892. https://doi.org/10.1007/s00449-012-0821-2.
Ravikumar, S., Ganesh, I., Maruthamuthu, M. K., & Hong, S. H. (2015). Engineering Escherichia coli to sense acidic amino acids by introduction of a chimeric two-component system. Korean Journal of Chemical Engineering, 32(10), 2073–2077. https://doi.org/10.1007/s11814-015-0024-z.
Utsumi, R., Brissette, R. E., Rampersaud, A., Forst, S. A., Oosawa, K., & Inouye, M. (1989). Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science, 245(4923), 1246–1249. https://doi.org/10.1126/science.2476847.
Ganesh, I., Ravikumar, S., Lee, S. H., Park, S. J., & Hong, S. H. (2013). Engineered fumarate sensing Escherichia coli based on novel chimeric two-component system. Journal of Biotechnology, 168(4), 560–566. https://doi.org/10.1016/j.jbiotec.2013.09.003.
Ganesh, I., Ravikumar, S., Yoo, I. K., & Hong, S. H. (2015). Construction of malate-sensing Escherichia coli by introduction of a novel chimeric two-component system. Bioprocess and Biosystems Engineering, 38(4), 797–804. https://doi.org/10.1007/s00449-014-1321-3.
Binder, S., Schendzielorz, G., Stabler, N., Krumbach, K., Hoffmann, K., Bott, M., & Eggeling, L. (2012). A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biology, 13(5), R40. https://doi.org/10.1186/gb-2012-13-5-r40.
Park, S. J., Kim, E. Y., Noh, W., Park, H. M., Oh, Y. H., Lee, S. H., Song, B. K., Jegal, J., & Lee, S. Y. (2013). Metabolic engineering of Escherichia coli for the production of 5-aminovalerate and glutarate as C5 platform chemicals. MetabolicEngineering, 16, 42–47. https://doi.org/10.1016/j.ymben.2012.11.011.
Park, S. J., Oh, Y. H., Noh, W., Kim, H. Y., Shin, J. H., Lee, E. G., Lee, S., David, Y., Baylon, M. G., Song, B. K., Jegal, J., Lee, S. Y., & Lee, S. H. (2014). High-level conversion of L-lysine into 5-aminovalerate that can be used for nylon 6,5 synthesis. Biotechnology Journal, 9(10), 1322–1328. https://doi.org/10.1002/biot.201400156.
Park, S. J., Jang, Y. A., Lee, H., Park, A. R., Yang, J. E., Shin, J., Oh, Y. H., Song, B. K., Jegal, J., Lee, S. H., & Lee, S. Y. (2013). Metabolic engineering of Ralstonia eutropha for the biosynthesis of 2-hydroxyacid-containing polyhydroxyalkanoates. Metabolic Engineering, 20, 20–28. https://doi.org/10.1016/j.ymben.2013.08.002.
Oh, Y. H., Eom, I. Y., Joo, J. C., Yu, J. H., Song, B. K., Lee, S. H., Hong, S. H., & Park, S. J. (2015). Recent advances in development of biomass pretreatment technologies used in biorefinery for the production of bio-based fuels, chemicals and polymers. Korean Journal of Chemical Engineering, 32(10), 1945–1959. https://doi.org/10.1007/s11814-015-0191-y.
Kim, H. S., Oh, Y. H., Jang, Y. A., Kang, K. H., David, Y., Yu, J. H., Song, B. K., Choi, J., Chang, Y. K., Joo, J. C., & Park, S. J. (2016). Recombinant Ralstonia eutropha engineered to utilize xylose and its use for the production of poly(3-hydroxybutyrate) from sunflower stalk hydrolysate solution. Microbial Cell Factory, 15, 95. https://doi.org/10.1186/s12934-016-0495-6.
Choi, S. Y., Park, S. J., Kim, W. J., Yang, J. E., Lee, H., Shin, J., & Lee, S. Y. (2016). One-step fermentative production of poly(lactate-co-glycolate) from carbohydrates in Escherichia coli. NatureBiotechnology, 34(4), 435–440. https://doi.org/10.1038/nbt.3485.
Joo, J. C., Oh, Y. H., Yu, J. H., Hyun, S. M., Khang, T. U., Kang, K. H., Song, B. K., Park, K., Oh, M. K., Lee, S. Y., & Park, S. J. (2017). Production of 5-aminovaleric acid in recombinant Corynebacterium glutamicum strains from a Miscanthus hydrolysate solution prepared by a newly developed Miscanthus hydrolysis process. Bioresource Technolology, 245(Pt B), 1692–1700. https://doi.org/10.1016/j.biortech.2017.05.131.
Yang, J. E., Kim, J. W., Oh, Y. H., Choi, S. Y., Lee, H., Park, A. R., Shin, J., Park, S. J., & Lee, S. Y. (2016). Biosynthesis of poly(2-hydroxyisovalerate-co-lactate) by metabolically engineered Escherichia coli. Biotechnology Journal, 11(12), 1572–1585. https://doi.org/10.1002/biot.201600420.
Chae, C. G., Kim, Y. J., Lee, S. J., Oh, Y. H., Yang, J. E., Joo, J. C., Kang, K. H., Jang, Y. A., Lee, H., Park, A. R., Song, B. K., Lee, S. Y., & Park, S. J. (2016). Biosynthesis of poly(2-hydroxybutyrate-co-lactate) in metabolically engineered Escherichia coli. Biotechnology and Bioprocess Engineering, 21, 169–174. 10.1007/s12257-015-0749-x, 1.
David, Y., Joo, J. C., Yang, J. E., Oh, Y. H., Lee, S. Y., & Park, S. J. (2017). Biosynthesis of 2-hydroxyacid-containing polyhydroxyalkanoates by employing butyryl-CoA transferases in metabolically engineered Escherichia coli. Biotechnology Journal, 12(11), 1–11. https://doi.org/10.1002/biot.201700116.
Wang, Y., Li, Q., Zheng, P., Guo, Y., Wang, L., Zhang, T., Sun, J., & Ma, Y. (2016). Evolving the l-lysine high-producing strain of Escherichia coli using a newly developed high-throughput screening method. Journal of Industrial Microbiology and Biotechnology, 43(9), 1227–1235. https://doi.org/10.1007/s10295-016-1803-1.
Pham, V. D., Lee, S. H., Park, S. J., & Hong, S. H. (2015). Production of gamma-aminobutyric acid from glucose by introduction of synthetic scaffolds between isocitrate dehydrogenase, glutamate synthase and glutamate decarboxylase in recombinant Escherichia coli. Journal of Biotechnology, 207, 52–57. https://doi.org/10.1016/j.jbiotec.2015.04.028.
Steil, L., Hoffmann, T., Budde, I., Volker, U., & Bremer, E. (2003). Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. Journal of Bacteriology, 185(21), 6358–6370. https://doi.org/10.1128/JB.185.21.6358-6370.2003.
Hadzhieva, T. (2007). Transcriptional activation and sensing properties of DegS-DegU: a two-component system involved in the osmotic regulation of Bacillus subtilis. PhD Thesis, Philipps University Marburg.
Mistry, J., & Finn, R. (2007). In N. H. Bergman (Ed.), Comparative genomics. Methods in molecular biology, vol 396: Pfam: a domain-centric method for analyzing proteins and proteomes (pp. 43–58). Totowa: Humana Press. https://doi.org/10.1007/978-1-59745-515-2_4.
Sambrook, J., & Russell, D. (2001). Molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
Epstein, W., & Kim, B. S. (1971). Potassium transport loci in Escherichia coli K-12. Journal of Bacteriology, 108(2), 639–644.
Nara, F., Matsuyama, S., Mizuno, T., & Mizushima, S. (1986). Molecular analysis of mutant ompR genes exhibiting different phenotypes as to osmoregulation of the ompF and ompC genes of Escherichia coli. Molecular Genetics and Genomics, 202(2), 194–199.
Ayyadurai, N., Neelamegam, R., Nagasundarapandian, S., Edwardraja, S., Park, H. S., Lee, S. J., Yoo, T. H., Yoon, H., & Lee, S. G. (2009). Importance of expression system in the production of unnatural recombinant proteins in Escherichia coli. Biotechnology and Bioprocess Engineering, 14(3), 257–265. https://doi.org/10.1007/s12257-009-0009-z.
Jin, T., & Inouye, M. (1993). Ligand binding to the receptor domain regulates the ratio of kinase to phosphatase activities of the signaling domain of the hybrid Escherichia coli transmembrane receptor, Taz1. Journal of Molecular Biology, 232(2), 484–492. https://doi.org/10.1006/jmbi.1993.1404.
Maeda, S., Takayanagi, K., Nishimura, Y., Maruyama, T., Sato, K., & Mizuno, T. (1991). Activation of the osmoregulated ompC gene by the OmpR protein in Escherichia coli: a study involving synthetic OmpR-binding sequences. Journal of Biochemistry, 110(3), 324–327. https://doi.org/10.1093/oxfordjournals.jbchem.a123579.
Levskaya, A., Chevalier, A. A., Tabor, J. J., Simpson, Z. B., Lavery, L. A., Levy, M., Davidson, E. A., Scouras, A., Ellington, A. D., Marcotte, E. M., & Voigt, C. A. (2005). Synthetic biology: engineering Escherichia coli to see light. Nature, 438(7067), 441–442. https://doi.org/10.1038/nature04405.
May, T., & Okabe, S. (2008). Escherichia coli harboring a natural IncF conjugative F plasmid develops complex mature biofilms by stimulating synthesis of colanic acid and Curli. Journal of Bacteriology, 190(22), 7479–7490. https://doi.org/10.1128/JB.00823-08.
Ohashi, K., Yamashino, T., & Mizuno, T. (2005). Molecular basis for promoter selectivity of the transcriptional activator OmpR of Escherichia coli: isolation of mutants that can activate the non-cognate kdpABC promoter. Journal of Biochemistry, 137(1), 51–59. https://doi.org/10.1093/jb/mvi006.
Rizk, S. S., Cuneo, M. J., & Hellinga, H. W. (2006). Identification of cognate ligands for the Escherichia coli phnD protein product and engineering of a reagentless fluorescent biosensor for phosphonates. Protein Science, 15(7), 1745–1751. https://doi.org/10.1110/ps.062135206.
Rogov, V. V., Rogova, N. Y., Bernhard, F., Koglin, A., Lohr, F., & Dotsch, V. (2006). A new structural domain in the Escherichia coli RcsC hybrid sensor kinase connects histidine kinase and phosphoreceiver domains. Journal of Molecular Biology, 364(1), 68–79. https://doi.org/10.1016/j.jmb.2006.07.052.
Diaz, E., & Prieto, M. A. (2000). Bacterial promoters triggering biodegradation of aromatic pollutants. Current Opinion in Biotechnolgy, 11(5), 467–475. https://doi.org/10.1016/S0958-1669(00)00126-9.
Georgellis, D., Kwon, O., & Lin, E. C. C. (2001). Quinones as the redox signal for the arc two-component system of bacteria. Science, 292(5525), 2314–2316. https://doi.org/10.1126/science.1059361.
Michalodimitrakis, K. M., Sourjik, V., & Serrano, L. (2005). Plasticity in amino acid sensing of the chimeric receptor Taz. Molecular Microbiology, 58(1), 257–266. https://doi.org/10.1111/j.1365-2958.2005.04821.x.
Ravikumar, S., Baylon, M. G., Park, S. J., & Choi, J. (2017). Engineered microbial biosensors based on bacterial two-component systems as synthetic biotechnology platforms in bioremediation and biorefinery. Microbial Cell Factories, 16, 62. https://doi.org/10.1186/s12934-017-0675-z.
Funding
This study was financially supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. NRF-2015R1A2A2A01004733), Golden Seed Project (213004-04-2-SBA30), Ministry of Agriculture, Ministry of Oceans and Fisheries, Korea Polar Research Institute grant (PE14070), and the Mid-Career Researcher Program through the NRF of Korea funded by the MSIT (NRF-2016R1A2B4008707).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic Supplementary Material
Fig. S1
(GIF 81 kb)
Rights and permissions
About this article
Cite this article
Ravikumar, S., David, Y., Park, S.J. et al. A Chimeric Two-Component Regulatory System-Based Escherichia coli Biosensor Engineered to Detect Glutamate. Appl Biochem Biotechnol 186, 335–349 (2018). https://doi.org/10.1007/s12010-018-2746-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-018-2746-y