Abstract
Invasive plants offer an interesting and unconventional source of protein and the considerable investment made towards their eradication can potentially be salvaged through their revalorisation. To identify viable sources, effective and high-throughput screening methods are required, as well as efficient procedures to isolate these components. Rigorous assessment of low-cost, high-throughput screening assays for total sugar, phenolics and protein was performed, and ninhydrin, Lever and Fast Blue assays were found to be most suitable owing to high reliability scores and false positive errors less than 1%. These assays were used to characterise invasive Scottish plants such as Gorse (Ulex europeans), Broom (Cystisus scoparius) and Fireweed (Chamaenerion angustifolium). Protein extraction (alkali-, heat- and enzyme assisted) were tested on these plants, and further purification (acid and ethanol precipitation, as well as ultrafiltration) procedures were tested on Gorse, based on protein recovery values. Cellulase treatment and ethanol precipitation gave the highest protein recovery (64.0 ± 0.5%) and purity (96.8 ± 0.1%) with Gorse. The amino acid profile of the purified protein revealed high levels of essential amino acids (34.8 ± 0.0%). Comparison of results with preceding literature revealed a strong association between amino acid profiles and overall protein recovery with the extraction method employed. The final purity of the protein concentrates was closely associated to the protein content of the initial plant mass. Leaf protein extraction technology can effectively raise crop harvest indices, revalorise underutilised plants and waste streams.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pressures exerted on agriculture require it to maintain profitability while ensuring adequate and sustainable food supply. The intensification strategies adopted to cope with demand are often at odds with environmental concerns, which provides scope for further investigation in unconventional and sustainable sources of nutrition [1]. However, mitigation strategies must work cohesively with the existing system as food supply chains function in tight cohesion and any attempts at rapid or extreme changes can be met with resistance or cause disruption in the system [2, 3]. Invasive plants competing with commercial crops in prime land is often a cause of hindrance to farmers, and the losses borne by them are quite significant. Bajželj et al. [4] detail the carbon flow through agricultural systems and estimate about 13% of cropland productivity being lost through unwanted plant intrusion (equating to about 1.0 Pg of biological carbon per annum). This is a cause of concern, particularly for subsistence farmers who may not have the resources to cope with such losses. Since low income countries tend to have agrarian economies, the impact of such losses is expected to be higher [5, 6]. Farmer distress is further amplified owing to the absence of compensation mechanisms and infrastructures to support them during crises [7].
The use of herbicides, herbivory and incineration are currently the dominant forms of plant removal. Incineration is the least satisfactory method of disposal given its contribution to pollution, the fire hazard it poses and the collateral loss of other flora and fauna, as witnessed in the recent fires in the Amazon basin [8] and Australia [9]. Herbicides such as glyphosate have received negative reports owing to their possible contribution towards the onset of non-Hodgkin’s lymphoma upon long-term exposure [10, 11]. Improper use of herbicides may also cause collateral loss of local flora, bioaccumulate through ecological food chains and contaminate ground water supplies. Herbivory employs domesticated ruminants to remove unwanted plant mass which may appear as satisfactory short-term means of biological revalorisation, but, certain wild plants contain anti-nutrients, which renders them unsuitable for animal feed [12, 13].
The concept of a circular economy provides a strong substratum upon which sustainable intensification strategies such as the revalorisation of unwanted plants can be realised, through the production of leaf protein concentrates (LPC). The concept and production of LPC is well established and its nutritional relevance to the human diet with respect to essential amino acid content has been extensively reported. However, despite the abundance of reports, adoption has been poor due to concerns of scalability [14], protein quality (particularly with respect to methionine [15, 16] and tryptophan [17, 18–20] content) and economic viability [21, 22]. Pioneers such as Pirie [23, 22] and Telek [24, 25] have investigated and enlisted numerous plant species pliable to leaf protein extraction and describe large-scale setups capable of efficient protein extraction through mechanical shearing. Fafunso et al. [26] demonstrated scalability and provided some insight to the economic feasibility of a large-scale commercial extraction unit, although final margins are highly dependent on market demand and subsidy policies. The work described here attempts to re-visit leaf protein extraction methods such as alkali, autoclave and enzyme assisted protein extraction methods through the lens of a circular and sustainable production setup to address the issue of food loss and farmer distress. We add data to the existing pool of literature on leaf protein extraction and look for overarching trends which may emerge across the various methods previously reported, which may help tailor process designs and allow optimal protein recovery and quality. The extracted leaf protein could be used in novel foods to aid replacing less sustainable animal products.
Scotland presents as an interesting case as globally recognised invasive plants [27–29] such as Gorse (Ulex europaeus), Broom (Cystisus scoparius) and Fireweed (Chamaenerion angustifolium) are native to the local Highlands. Having gone feral through improper horticultural management when introduced in foreign lands [30], the characterisation of these plants may now serve to realise their nutritional potential and allow for their rapid removal and consumption in non-native lands, while promoting their conservation in their native niches. We investigated some of the food production trends in Scotland and assessed the potential of such underutilised plants to complement the existing supply chain by introducing diversity in nutritional sources and relieving the burden currently placed on animal husbandry for high quality protein production.
Materials and methods
All chemicals and kits were purchased from Merck (Darmstadt, Germany) and used without further modification unless stated otherwise. All experiments and measurements were performed in triplicate. The colorimetric assays were carried out in 96-well plate format and incubated with film cover (Greiner Bio-One; Kremsmuenster, Austria). Absorption values were obtained using a SpectraMax 190 (Molecular Devices; San Jose, USA). Water used in the experiments was from Milli-Q®.
Scotland agriculture data
Scottish gross domestic product (GDP) and carbon emission data were obtained from the Scottish Government for 2018 [31, 32]. GDP value of “Mining”, “Construction” and “Manufacturing” were grouped into “Industrial Production”. In the dataset, carbon emission for “Business” were included in values for “Exports”.
Food and feed production data was obtained from HMS Revenue services [33]. In the cases of generalised groups such as “cereals” or “other fruits”, the average known values of amino acids were considered. Information on land capability was obtained from James Hutton Institute [34]. When a given area is collectively allocated for multiple crops, equal land distribution per crop was assumed. Data on food amino acid content was obtained from the USFDA [35].
Leaf protein extraction data
Previous research on leaf protein extraction were obtained from three databases: namely, Google Scholar, Web of Science and Scopus. The search terms used were “Leaf” AND “Protein” AND (“Concentrate” OR “Extraction”) with a time frame between 1970 to May 2020. Patents were disregarded from the search as they aimed to describe overall processes and failed to accurately define protein recovery and purity values of the concentrates. In the data analysis, “Extraction” referred to the method adopted to obtain protein from the plant source. “Purification” referred to the method adopted to enrich protein from the extract obtained in the preceding extraction step. “Process” referred to the combination of Extraction and Purification methods adopted by the author to produce a protein concentrate.
Plant sample collection
Plant sample collection and aqueous extraction was performed as previously described in Iyer et al. [6]. Briefly, Gorse (Ulex europaeus) and Broom (Cystisus scoparius) were collected from GPS co-ordinate: 57.257, \(-2.483\) and Fireweed (Chamaenerion angustifolium) from GPS co-ordinates: 57.157, \(-2.086\). Sampling was performed based on guidelines laid down by British Columbia Ministry of Forests Handbook [36]. The leaves were separated from the plant stalks, freeze-dried (Labconco, UK), milled (Centramex, USA), and stored at \({4}\,^\circ{\mathrm{C}}\) under vacuum for a period of up to two months prior to analysis or extraction.
Sample preparation
Freeze-dried leaf samples of each plant were stirred in phosphate buffered solution, (PBS; 10 \(\,\mathrm{mM}\), pH 7.5) (1:10 :: plant: buffer) for 20 min on a magnetic stirrer at \({32}\,^\circ {\mathrm{C}}\), then sonicated in a water-bath for 10 min and then stirred for 10 min. Samples were then centrifuged at 14,000×g for 15 min at \(4\,^\circ \mathrm{C}\). Supernatant and retentate were separated, frozen at \({-70}\,^\circ {\mathrm{C}}\) and freeze-dried. Samples obtained were weighed and characterised with the methods described below. The supernatant is herein referred to, as the ‘First Extract’. The retentate was subjected to further experimental extraction procedures described below and is herein referred to as Experimental Retentate (ER). Each extraction method (described in the sections below) was performed using 0.7g of ER for each replicate.
Screening methods
Protocols are described in the following subsections. All incubation steps were performed in the dark so as to avoid interference due to photo-oxidation.
Standards
Standard solutions were prepared for protein, sugar, and phenolic estimation. Bovine serum albumin (BSA) was prepared in phosphate buffered solution (PBS; \({0.2}\,\mathrm{mM}\)), while for phenolics and sugars, gallic acid and glucose were dissolved in water. All standards were prepared at concentrations of 1, 0.75, 0.5, 0.25, 0.125 and 0.0625 mg/mL and for each assay, measurements were made in triplicate and were repeated on three separate days. For the Lever assay, calibration curves were also generated using xylose and uronic acid.
Protein estimation methods
Ninhydrin assay: Ninhydrin measures proteins by estimating total amino acid content of a hydrolysed sample as previously described by Harding et al. [37]. Procedure and details of the method adapted for a 96-well plate setup has been described previously by Iyer et al. [6]. Briefly, sample or standard (\({200}\,{\upmu }\!\mathrm{L}\)) was incubated with HCl (anærobic, 6 M with phenol 2% w/v) at \({110}\,^\circ \mathrm{C}\) for 14 h in heating blocks. The digesta (\({10}\,{\upmu }\mathrm{L}\)) was dried in a 96-well plate at \({60}\,^\circ \mathrm{C}\) which was re-dissolved in water (\({100}\,{\upmu }\mathrm{L}\)). Ninhydrin solution (2% w/v , Sigma-Aldrich, Missouri, USA, \({75}\,{\upmu }\!\mathrm{L}\)), was added and incubated at \({60}\,^\circ \mathrm{C}\) for 60 min, and the absorbance was measured at 570 nm.
Bradford assay: Assay was based on the work by Bradford [38]. Bradford reagent was purchased from Merck and procedure was followed according to the manufacturer’s instruction. The reagent (\({250}\,{\upmu }\!\mathrm{L}\)) was mixed with sample or standard (\({5}\,{\upmu }\!\mathrm{L}\)) on a shaker for 30 s. The analytes were allowed to rest for a further 5 min at room temperature. Absorbance was measured at 595 nm.
BCA (Bicinchoninic acid) assay: This assay was based on the work by Mallia et al. [39]. The BCA assay kit was purchased from Thermo-Ficher, (Massachusetts, USA) and was performed in accordance to the manufacturer’s instructions. Standard (\({10}\,{\upmu }\!\mathrm{L}\)) was incubated with \({200}\,{\upmu }\!\mathrm{L}\) of BCA working reagent in a 96-well plate for 30 s on a shaker and then allowed to rest at 37 \(^\circ \mathrm{C}\) for 30 min. Absorbance was measured 562 nm.
Pierce 660: This assay was based on the work by Antharavally et al. [40]. Pierce 660 kit was purchased from Fischer Scientific, (Loughborough, UK) and was carried out according to the manufacturer’s instructions. Standard or sample (\({10}\,{\upmu }\!\mathrm{L}\)) was mixed with Pierce 660 reagent (\({150}\,{\upmu }\!\mathrm{L}\)) with a shaker for 30 s. Analytes were incubated for 5 min at room temperature, and absorbance was measured at 660 nm.
Biuret assay: Reagent for the biuret test was prepared following the recipe described by Potty [41]. Standard or sample (\({150}\,{\upmu }\!\mathrm{L}\)) was added to \({50}\,{\upmu }\!\mathrm{L}\) of reagent in a 96-well plate and incubated on a shaker for 30 s. Samples were further incubated without shaking for 20 min at room temperature and the absorbance was measured at 540 nm.
Lowry assay: Standard or sample (\({50}\,{\upmu }\!\mathrm{L}\)) was mixed with biuret reagent (\({50}\,{\upmu }\!\mathrm{L}\)) and incubated for 10 min at room temperature in a 96-well plate on a shaker. Folin-Ciocâlteu reagent (Sigma-Aldrich; Dorset, U.K; 1 M; \({200}\,{\upmu }\!\mathrm{L}\)) was added and incubated further on the shaker for 30 s, after which the samples were incubated at 55 \(^\circ \mathrm{C}\) for 5 min. Absorbance was measured at 740 nm.
Phenolic estimation methods
Folin–Ciocâlteu assay: This method was carried out as described in Lester et al. [42]. Standard or sample (\({25}\,{\upmu }\!\mathrm{L}\)) was mixed with Folin–Ciocâlteu reagent (0.33 M; \({25}\,{\upmu }\!\mathrm{L}\)), to which water (\({200}\,{\upmu }\!\mathrm{L}\)) was added and incubated on a shaker for 5 min. To the samples, sodium carbonate solution (Na2CO3; 25% w/v ; \({25}\,{\upmu }\!\mathrm{L}\)) was added and incubated for a further 60 min in the dark. Absorbance was measured at 725 nm.
Fast Blue: This method was based on the work described by Lester et al. [42]. The procedure and reagent composition has been described previously by Iyer et al. [6]. Briefly, the Fast Blue dye (o-dianisidine bis(diazotized) zinc double salt, 0.1% w/v ) was added to sample or standard (\({100}\,{\upmu }\!\mathrm{L}\)) and incubated on a shaker for 30 s. To this, sodium hydroxide (NaOH; \({2}\,{\upmu }\!\mathrm{L}\); 5% w/v ) was added and incubated for 90 min after which, absorbance was measured at 420 nm.
Prussian Blue: Prussian Blue assay was carried out as described by Puyeo et al. [43]. Standard or sample (\({10}\,{\upmu }\!\mathrm{L}\)) was raised to \({100}\,{\upmu }\!\mathrm{L}\) with water to which, FeCl2·6H2O (0.5 mM; \({100}\,{\upmu }\!\mathrm{L}\)) solution prepared in HCl (10 mM) was added and allowed to rest for 5 min at ambient conditions. Prussian Blue dye solution (K3(Fe(CN)6), 0.5 mM, \({100}\,{\upmu }\!\mathrm{L}\)) was added to the sample and kept on a shaker for 30 s. The samples were incubated for 15 min and absorbance was measured at 725 nm.
Sugar estimation methods
Lever assay: This assay was based on the work described previously by Lever [44, 45]. Reagent composition and procedure has been described previously by Iyer et al. [6]. Briefly, the Lever reagent comprised of 4-hydroxybenzoic acid hydrazide (PAHBAH; 0.76% w/w), bismuth (III) nitrate pentahydrate (Bi(NO3)3·5H2O, 0.48% w/v), potassium sodium tartrate (0.28% w/v), NaOH (2\(\%\) w/v) in water. Sample or standard (\({5}\,{\upmu }\!\mathrm{L}\)) was incubated with Lever reagent (\({200}\,{\upmu }\!\mathrm{L}\)) at 70 \(^\circ \mathrm{C}\) for 30 min. The sample was then allowed to cool for 10 min and absorbance was checked at 415 nm.
Anthrone assay: Anthrone assay was carried out based on the work described previously [46]. Sample or standard (\({50}\,{\upmu }\!\mathrm{L}\)) was incubated with of anthrone (0.1\(\%\) w/v, \({150}\,{\upmu }\!\mathrm{L}\)) solution in conc. H\(_{2}\)SO\(_{4}\) (98% v/v) at 4 \(^\circ \mathrm{C}\) for 5 min. The sample was then incubated at 100 \(^\circ \mathrm{C}\) for 10 min and then allowed to cool for 5 min. Absorbance was checked at 620 nm.
Phenol-sulphuric acid: This assay was carried out based on the work described previously [47, 48]. Sample volume of \({50}\,{\upmu }\!\mathrm{L}\) was incubated with phenol (5% w/v) solution in sulphuric acid (H2SO4, 98% w/v) at 4 \(^\circ \mathrm{C}\) for 5 min. The sample was then incubated at 100 \(^\circ \mathrm{C}\) for 10 min and then allowed to cool for 5 min. Absorbance was checked at 490 nm.
Measurement of assay suitability
The suitability of assays was established based on their reliability and selectivity. Reliability (RL) was calculated as the root mean square (RMS) value of repeatability (Rt), reproducibility (Ro), and accuracy (Ra).
Repeatability (Rt) represented the percent consistency of a result obtained for a triplicate measurement on a given day. It was calculated by the formula \(100-\sigma ^{2}_{t\%}\), where \(\sigma ^{2}_{t\%}\) was the percent variance obtained across the results obtained in the triplicate measurement.
Reproducibility (Ro) represented the percent consistency of a result obtained for a measurement repeated across three different days. It was calculated by the formula \(100-\sigma ^{2}_{o\%}\), where \(\sigma ^{2}_{o\%}\) was the percent variance obtained across the results obtained in the triplicate measurement.
Accuracy (Ra) represented the percentage ratio \(\frac{C_{m}}{C_{w}}\) of measured concentration (Cm) of a standard compound using the assay, against the known weighed concentration (Cw).
Thus, the final Reliability was calculated by the formula: \(R_{L} = \sqrt{\frac{(100-\sigma ^{2}_{t\%})+(100-\sigma ^{2}_{o\%})+(\frac{C_{m}}{C_{w}} \times 100)}{3}}\).
Selectivity was a measure of the assays ability to faithfully estimate its target compound. First, assay was performed on standard compounds used for estimation of the other two compound groups, which in this scenario would behave as “contaminants”. False positive response was measured as the average percentage absorbance ratio of contaminant to relevant standard at the same concentration. Assays with false positive values \(<1\)% were further tested against a bovine serum albumin (BSA) + glucose + gallic acid mixture to check for changes relative to their calibration profile.
Extraction conditions
Enzyme Treatment: Three saccharolytic enzymes; cellulase (EC 3.2.1.4), xylanase (EC 3.2.1.8), pectinase (EC 3.2.1.15), a lignolytic enzyme; laccase (EC 1.10.3.2), and a proteolytic enzyme; papain (E.C 3.4.22.2) were used at concentrations of 0.3, 0.6 or 1% (w/w E.R). Two additive combinations of cellulase + xylanase + pectinase (saccharase) and cellulase + xylanase + pectinase + laccase (mixture) were also investigated.
Saccharolytic and lignolytic experiments were performed in sodium citrate buffer (50 mM; pH 4.5; 15 mL) at 40 \(^\circ \mathrm{C}\), while the proteolytic experiment was carried out at 47 \(^\circ \mathrm{C}\) with pH 6.5. E.R suspended in buffer without any enzyme was used as negative control. Buffer with enzyme, but without E.R. was used as a control to measure the protein contribution from the enzyme. Samples were taken at 0, 0.5, 1, 2, 3, 4 and 24 h time points and centrifuged at 14,000\(\times \)g for 15 min at 4 \(^\circ \mathrm{C}\). Supernatants were collected as fraction of interest for estimation of protein, sugar and phenolics.
Alkali treatment: Extraction was performed as described by Zhang et al. [49, 50]. The sample was stirred in 15 mL of NaOH (0.4 M) at 95 \(^\circ \mathrm{C}\) for 4 h. Negative control condition was water at 21 \(^\circ \mathrm{C}\) (ambient temperature) for 4 h. Precipitation was carried out by lowering the pH to 3.5 using HCl solution (1 M). Samples were centrifuged at 14,000\(\times \) g for 30 min at 15 \(^\circ \mathrm{C}\). The precipitate was collected for protein, sugar, and phenolic estimations.
Heat treatment: ER was subjected to heat treatment using a medical autoclave (Prestige, UK) using 15 mL water. The temperature was raised to 121 \(^\circ \mathrm{C}\) and held for 15 min followed by a 2 h cooling period. The sample was vigorously vortexed and centrifuged at 14,000\(\times \)g for 15 min at 4 \(^\circ \mathrm{C}\). Negative control condition was 21 \(^\circ \mathrm{C}\) (ambient temperature) in water. The supernatant was collected assayed for protein, sugar and phenolics.
Amino acid estimation
Hydrolysis medium for amino acid estimation comprised of HCl (anærobic, 6 M) doped with phenol (3% w/v) and tryptamine (0.2% w/v). Samples were first subjected to microwave assisted digestion (MARS 6, CEM, USA). They were loaded into Teflon®tubes (final capacity 20 mL) containing 10 mL of hydrolysis medium. The heating regime of the microwave involved raising the temperature to 155 \(^\circ \mathrm{C}\) over a period of 15 min and holding it for a further 5 min after which the heating is stopped and the samples are allowed to cool over a period of 4 h. Amino acids were profiled and quantified using GC-MS with 13C amino acids as internal standards as described previously by Calder et al. [51] with some modifications.
Internal 13C standards were added to the digestates (1:1) and purified by using cation exchange chromatogrpahy (AG50W resin, BioRad). Resin was washed twice with water (2 mL) and eluted using NH4OH (2 mL), followed by water again (1 mL). The eluates were collected, freeze dried and resuspended in HCl (0.1 M, 0.35 mL) and dried at 90 \(^\circ \mathrm{C}\) under N2. Finally, the samples were derivatised using MTBSTFA/DMF (1:1) at 120 \(^\circ \mathrm{C}\) for 20 min.
GC/MS analysis was performed on a HP 5989A MS Engine coupled to a HP 5890 gas chromatograph (Hewlett Packard, Manchester, UK) at injection temperature of 280 \(^\circ \mathrm{C}\) and the interface line at 250 \(^\circ \mathrm{C}\). Amino acid separation occurred across a 30 m \(\times \) 0.25 mm \(\times \) 0.25 mm SE-30 CB capillary column (Alltech, Carnforth, UK) at 160 \(^\circ \mathrm{C}\) for 5 min then then rasied at a rate of 15 \(^\circ \)C/min to 270 \(^\circ \)C over 8 min. Injections (1 mL) were made with a 40:1 split using helium (at 11 psi head space) as the carrier gas. The capillary column was connected directly to the ion source of the mass spectrometer, which operated in the EI mode with an electron energy of 70 eV, emission current of 300 mA and a source temperature 200 \(^\circ \)C. Amino acid and internal standard fragment ions were monitored with a 30 ms dwell time for each ion, 20 ms inter-channel delay under SIM conditions.
Non-starch polysaccharide (NSP) determination
Non-starch polysaccharides were quantified as described previously [52, 53] with no modifications. Briefly, plant samples were hydrolysed in H2SO4 (7 M) at 100 \(^\circ \mathrm{C}\) for 1h. Monosaccharides were analysed by Gas Chromatography with Flame Ionised Detection (GC-FID) using inositol as an internal standard. Charged residues such as uronic acids was measured colorimetrically using glucuronic acid as standard [54].
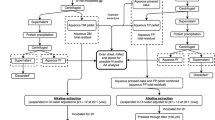
Scaled-up extraction
The extraction was scaled up using 20 g of freeze milled Gorse samples suspended in of PBS (10 mM; pH 7.5; 200 mL) and stirred for 20 min at 32 \(^\circ \mathrm{C}\). Sample was centrifuged at 3000\(\times \)g for 15 min and the supernatant was recovered. Retentate was resuspended in sodium citrate buffer (50 mM; pH 4.5; 200 mL), at 40 \(^\circ \mathrm{C}\) and incubated with cellulase (1% w/w of dry retentate) for 2 h. Samples were centrifuged at 3000\(\times \)g for 15 min and the supernatant recovered. Supernatant from the preliminary PBS extract and the following enzyme treatment were pooled and used for further purification experiments. This will hence be referred to as Gorse Scaled Extract (GSE).
Purification methods
Ethanol precipitation: GSE (20 mL) was incubated with ethanol (7 mL) at 4 \(^\circ \mathrm{C}\) for 60 min and centrifuged at 8000\(\times \) g for 15 min. The supernatant was evaporated at 40 \(^\circ \mathrm{C}\) under vacuum using a rotary evaporator at 20 Torr until volume was reduced by approximately 60%. The remaining 40% volume was freeze-dried along with the precipitate and this was retained for amino acid profiling. Ethanol purity was measured using a floating densitometer (HB Instruments, Pennsylvania, US).
Acid precipitation: GSE (20 mL) was adjusted to pH 2 using HCl (2 M) over a period of 2 h under stirring at ambient temperature and allowed to rest at 4 \(^\circ \mathrm{C}\) for 60 min. Samples were centrifuged at 8000\(\times \)g for 15 min and supernatant and precipitate were separated. Supernatant was neutralised using NaOH (2 M) and freeze-dried and analysed for protein and amino acids.
Ultrafiltration: GSE (80 mL) was subjected to ultrafiltration using benchtop Sartorius ultrafiltration unit (Vivaflow 50R Hydrosart; Epsom, UK) with a molecular weight cut off (MWCO) of 10 kDa. Back pressure was at 1 bar when flow rate was adjusted to 0.6 mL/min. Volume was kept constant by feeding PBS (pH 7.5, 10 mM) until a passage of 240 mL after which time the sample volume was dropped to 40 mL. The filtrate and retentate were separated and freeze dried.
Statistics and data analysis
Statistical analysis was performed using R (Version 3.6.1) [55] and RStudio (Version 1.2.1335). Principal Component Analysis (PCA-Univariate scaled) was performed using packages ‘factoextra’ [56] with additional packages: ‘ggplot2’ [57] and ‘corrplot’ [58]. Significance was tested using one-way ANOVA with ad hoc Tukey HSD test unless stated otherwise. Statistical results are expressed as F(degrees of freedom, residuals) = F-value; p-value as suggested by Field et al. [59].
Results and discussion
Across the abundant literature available on leaf protein extraction, numerous assays, extraction and purification methods have been reported which we attempt to summarise, assess and apply towards the revalorising invasive plant species as an alternate protein source. Accurate and reliable calculation of mass balance is pivotal to assessing the efficacy of leaf protein extraction and purification strategies.
Assay performance
Among the twelve colorimetric assays tested for reliability, six were designed for protein estimation, three for phenolic estimation and three for sugar estimation. Of these, only seven were found to measure standards consistently with a reproducibility greater than 90%, and repeatability and accuracy greater than 95%, namely, ninhydrin, Bradford, and BCA for protein estimation, Folin–Ciocâlteu, Fast Blue and Prussian Blue for phenolics estimation and the Lever assay for sugar estimation as shown in Supplementary Table 1 below. Of these seven methods, only three were able give a false positive response of less than or equal to 1%, namely, ninhydrin for protein, Fast Blue for phenolics and Lever for sugar, as detailed in Supplementary Table 2. Furthermore, ninhydrin, Fast Blue, and Lever assays were performed on a BSA-glucose–gallic acid mixture of known concentration to check for inaccuracies generated in a composite sample. However, no difference in recoveries were found and standard curves generated across multiple concentrates showed no significant difference using one-way ANOVA with a 99% confidence interval as shown in Figure 1 and detailed in Supplementary Table 3.
Assay sensitivity was determined using Limit of Blank (LoB), Limit of Detection (LoD) and Limit of Quantification (LoQ). Based on slope values from Supplementary Table 3 and the low LoQ values in Table 1, ninhydrin appears to be a very sensitive method for protein estimation.
The sensitivity for Fast Blue and Lever assays were found to be similar. The LoQ was lower for the Lever assay due to the high background reading, which was likely due to oxidation catalysed by the bismuth ions on PAHBAH [45]. However this helps improve assay reliability by decreasing measurement variance [44].
Extraction treatments
Gorse, Broom and Fireweed were subjected to extraction using alkali, autoclave and enzyme assisted extraction methods to compare protein recovery. A summary of protein recovery across the different extraction stages is shown in Table 2. Most of the recoverable protein was present in the first extract. Protein content of ER from Gorse was 8.4 ± 1.0% (w/w), of Broom was 10.1 ± 0.0% (w/w) and Fireweed was 17.8 ± 0.0% (w/w).
With enzyme treatment, highest recoveries were obtained at enzyme concentrations at 1% (w/w ER). Protein recovery from individual enzyme conditions is depicted in Supplementary Figure 1. For each plant, there appeared to be a primary cell-wall constituent homo-polymer from which much of the bound protein was recovered.
In the case of Gorse and Broom, the action of the single enzyme cellulase was comparable to the mixed enzyme conditions. Across these two plant candidates, the mean protein recovery is 83.5% which is higher than previously published recoveries using this method (discussed further in the following Sect. 3.5), with the highest at 61.99% reported by [60]. In the case of Fireweed however, xylanase was comparable to the mixed enzyme conditions suggesting a major carbohydrate polymer to which proteins are bound in the retentate. Protein recovered due to enzyme action (“Effective” in Table 2) was found to be highest for Broom where 16.6 ± 2.3% (w/w) total protein was recovered due to cellulase action. Given the ease of working with single enzymes and their economy, further focus was made on the digestion profiles using these conditions.
Protein and sugar recovery profiles for Gorse, Broom and Fireweed at enzyme concentration of 1% (w/w experimental retentate). Broom and Gorse profiles are following cellulase treatment while the Fireweed profile follows xylanase treatment. Dotted vertical line in panels Protein and Ratio represent the time point at which protein recovery rate begins to plateau
Xylose recovery from Fireweed was almost 80% within the first 30 min of incubation with an end point at three hours, where subsequent recovery measurements were statistically similar. On the other hand, cellulase treatment for Gorse and Broom showed low sugar recovery despite their high concentration relative to bound protein. While glucose recovery from Broom was relatively modest at 30.8 ± 1.3% following a four-hour treatment, recovery from Gorse was approximately half at 16.3 ± 0.9%. For Broom and Fireweed, recovery begins to plateau after two hours of enzyme treatment (Fig. 2, Panel: ‘Sugar’). For Gorse however, a marginal rise in protein levels is observed until 24 h, suggesting the slow release of protein into the medium. The effect of cell wall digestion on protein recovery may be visualised through values expressing the ratio of % protein recovery to % sugar recovery, as shown in Figure 2, Panel: ‘Ratio’. A rapid increase in the recovery ratio would indicate protein release owing to enzyme action, followed by either a plateau or a decline once an equilibrium or excess sugar release is achieved. This is likely to be due to continued cell-wall digestion and protein exhaustion.
The highest recovery ratio was observed for Broom which suggests a strong association between protein and cellulose. In Figure 2, we observe a peak recovery ratio at two hours suggesting that most of the protein had effectively been released by that point. This corresponds to a glucose recovery of 24.7 ± 1.7% from cellulose digestion and a protein recovery of 12.1 ± 3.8% effectively from enzyme action. Effective protein recovery at the two-hour time point is statistically similar to the values at the 4 h and 24 h time points using one-way ANOVA at 95% confidence interval. Glucose recovered between two and three hours is about 4.6 ± 2.2% and the overall rate appears to decrease compared to the first two hours of incubation. This suggests that further protein recovery may not be dependent on enzyme action, but rather through passive diffusion.
In the case of Gorse, enzyme mediated protein release follows a similar trend to Broom, with recoverable bound protein was released only after two hours, when about 14.5 ± 0.5% of the total cellulose content was digested. Most of the bound protein appears to have been recovered through the digestion of 14-20% of the cellulose substrate in Gorse and Broom. Incomplete digestion of the cell wall despite high enzyme concentrations over a 24 h period suggests complex and resilient structures between various cell-wall components [61, 62, 63, 64]. Lowest effect of enzyme action on protein recovery was observed for Fireweed despite complete digestion of xylose. As Table 2 indicates, this result is comparable to other enzyme combinations despite wider substrate digestion suggesting the protein may either be insoluble or bound to locations other than the primary cell wall.
Extraction under alkaline (1 M to 0.4 M NaOH) conditions is a well established method of protein recovery [65]. Across the plants, the mean protein recovery was 73.96% which was comparable to previous publications by El-Sayed et al. [66], and Moure [67]. However, tryptophan and sulphur containing compounds are thought to be compromised during the neutralization step which lowers the nutritional value of the protein [20]. Autoclave treatment was employed as a means of exerting heat and physical pressure to lyse the plant cell walls to release bound proteins. Encouraging results were observed for Fireweed, where final recoveries were significantly higher compared to xylanase treatment (Table 2). Autoclave treatment of Broom gave modest results where final recoveries were marginally superior to alkali treatment and statistically similar to enzyme treatment. Autoclave treatment on Gorse appeared to have a detrimental effect on the overall protein recovery, with the protein recovered from the control samples being higher compared to autoclave treatment. The loss in recovery may be attributed to the coagulation and tighter association post autoclave treatment resulting in lowered protein recoveries. Gorse and Broom belong to the same phylogenic family (Fabaceae) and the bulk of the protein expressed was not expected to be significantly different from each other. Nonetheless, the highly modified thorny morphology of Gorse leaves may render these more resistant to elevated heat and pressure and allowing for protein coagulation within the structure rather than solubilising them into the surrounding medium.
These results appear to suggest that the effectiveness of any intervention is dependent on its ability to rupture the cell wall structure and release the bound protein. Table 3 below shows the protein and NSP content of the investigated plants. Gorse and Broom shared similar protein content while the NSP profile was different across all plants.
Significance was tested using one-way ANOVA at 95% confidence interval along with posthoc Tukey HSD. Values with the same suffix have statistically similar results.
In line with previous literature, the performance of an extraction method appeared to be specific for a given plant species. Alkali treatment was very effective for recovering proteins from Fireweed, while autoclave and enzyme treatment were equally suitable for Broom. In the case of Gorse, the most effective method was cellulase treatment, but the increase in recovery was marginal. Autoclave treatment brought about a decrease in the overall recovery. Among the plants investigated, the highest protein recovery was observed for Gorse with the bulk obtained through passive dissolution. Any further treatment lead to either a marginal increase or a significant decrease in protein recovery. For Gorse 76.5 ± 0.9% of the total protein was recovered through passive buffer extraction (Table 2), making it a suitable plant candidate for obtaining protein with minimal intervention. In the large scale cellulase treatment of Gorse, final recovery was 88.0 ± 3.0%. This result is statistically similar (F(1,11)=3.79, p=0.08) to the corresponding condition in Table 2. Total soluble sugar was 5.1 ± 0.3% (w/w dry plant mass) and total phenolic content was found to be 2.9 ± 0.3% (w/w dry plant mass).
Purification method comparison and carbon efficiency
Three protein enrichment methods were employed to refine protein in the Gorse Scaled Extract (GSE), one based on ultrafiltration and two based on precipitation using either acid or ethanol.
Recoveries were calculated relative to protein present in the scaled-up Gorse extract. ‘Protein’ refers to precipitate for acid and ethanol precipitation and the retentate in ultrafiltration. Among the methods tested, the highest recovery and purity was obtained for ethanol precipitation and the overall protein recovery was 56.32% (Table 4). Only 79.0 ± 8.0% ethanol used for precipitation was recovered through evaporation. Based on the protein recoveries obtained, ethanol consumption would amount to 11.2 L/kg leaf protein. Conservative carbon footprint associated to ethanol production is reported to be approximately 0.4 kg/L [68, 69]; although depending on the production method used, the value ranges from 1.3 to 0.2 kg/L [69]. The emissions associated to solvent use in production of 1 kg leaf protein would come to 4.5 kg CO\(_{2}\) eq./kg leaf protein.
The upper limit of carbon footprint can be determined through a pessimistic estimate of 1.3 kg CO\(_{2}\) eq. emission/L ethanol without any solvent recovery. The ethanol consumption would amount to 53.4 L/kg leaf protein resulting in a carbon footprint of 69.5 kg CO\(_{2}\) eq/kg leaf protein which is still lower than the emissions associated to protein obtained from animal husbandry as mentioned in the paragraphs below.
Amino acid profile of gorse protein extract
Amino acid composition of the extracts is given in Table 5 below.
The protein quality recovered in the precipitate was superior compared to the residual soluble protein in the supernatant based on the total essential amino acid content, which was three times higher while methionine was undetectable in the residual supernatant protein. Only the branched chain amino acids (BCAA), valine and isoleucine levels were comparable.
Leaf protein recovery and purity
A literature search on previous publications detailing leaf protein extraction and purification methods using Scopus, Web of Science and Google Scholar yielded 373 relevant hits of which only 40 gave sufficient details on mass balance and the protein content of the leaf mass. Since most publications reported extraction processes with more than one plant, in all, 75 cases of LPC were summarised. Alkali (n=29) and juicing (n=23) were the most widely employed extraction methods, while acid precipitation was the popular purification method (n=27). A summary of the protein content, recovery, protein purity, extraction and purification method employed by preceding literature is tabulated in Supplementary Table 4.
To obtain a clearer understanding of the effect of each stage in LPC production, reports of purity and recovery were first filtered using statistical package dplyr in R, according instances where extraction methods without subsequent purification steps were reported (Fig. 3).
One-way ANOVA found significant differences in recovery values across different extraction methods. The posthoc Tukey HSD test revealed significant differences in recoveries obtained from juicing and autoclave treatment, and juicing and enzyme treatment. The lack of resolution between alkali and juicing methods despite the clear difference in their respective average recovery values may have risen from the large variation across reported results. While the number of data points available for alkali and juicing were somewhat similar (seven [70, 71, 72, 49] and five [73, 74, 75] respectively), reports on juicing were primarily on Ryegrass and Lucerne, while the plants used in alkali treatment were more diverse.
The graph presented in Figure 4 and 5, represent average recovery and purity values which include instances where extraction and purification stages were used in combination. In Figure 4, where the values were grouped according to extraction method, one-way ANOVA, and posthoc Tukey HSD test revealed significance trends similar to those observed in Figure 3 with autoclave and enzyme recovery values being significantly greater than juicing. The purity values obtained across LPCs could not be statistically distinguished across extraction methods, except for autoclaving which was significantly lower.
Furthermore, ANOVA for purity expressed as an interaction between extraction and purification stages appeared to show significance (F(8,52)=3.15, p=0.005). Predictably, posthoc Tukey HSD revealed the purity value of Autoclave treatment with no purification step being significantly lower than alkali extraction-acid precipitation (p=0.006), enzyme extraction-acid precipitation (p=0.03). Lack of statistical distinction among other processes may have risen from the high variance (consequently high RMS error), owing to factors not considered in the model such as protein estimation errors, matrix effect of plant material and lack of data on other combinations of extraction and purification methods such as ultrasound with acid precipitation or autoclave with centrifugation. Lastly, one-way ANOVA was performed to assess association between initial protein content and the final recovery and purity of the protein concentrate. No significant association was found between protein recovery and initial protein content. Significant association (F(1,70)=12.91, p<0.001) was found between initial protein content of plant and the protein concentrate purity. Average values of initial protein content of plant material are shown in Supplementary Table 5.
These results appear to reflect a strong relationship between extraction method and the final protein recovered in the LPC, while the final purity of the concentrate is related to the initial protein content of the plant material used. This further explains the lack of association between purity values and purification methods as most purification methods rely on the relative difference in solubility between contaminant and protein and the trend appears to suggest a limited resolving power among these reported methods. Based on the available mass balance data, only heat (autoclave) and enzyme-based extractions showed significant difference in recovery compared to juicing while among purification methods, only acid and solvent based precipitation gave LPC purities significantly higher than untreated leaf extract. The analysis thus suggests that final protein recovered in the LPC was a function of the extraction process, while its purity was a function of the initial protein content in the leaf sample.
Effect of treatment on amino acid profile
Amino acid profile of the experimental precipitate generated from this work was compared to previously reported values as shown in Supplementary Table 6. Principal component analysis (PCA, univariate scaled) is shown in Figure 6 which demonstrates effects of processing on LPC amino acid profiles. A trend emerges where processes with juicing as part of the extraction process are found to appear in the lower two quadrants as whilst other extraction methods are mostly confined to the upper two quadrants.
PCA (univariate-scaled) plot of purity and amino acid profile of protein concentrates in previous literature marked according to Process. The ellipse demarcates 95% confidence interval around group means. Standard refers to the purification of Rubisco. The numbers represent the publication where the work was described, and corresponding details can be found in Supplementary Table 6
This trend appears equally distinct in Figure 7, which explores protein extraction where juicing is confined only to the lower two quadrants, while the other extraction methods are found in the upper two quadrants. Rubisco, which forms the bulk of soluble protein was expected to be recovered unaltered through the juicing process.
PCA (univariate scaled) plot of purity and amino acid profile of protein concentrates in previous literature marked according to extraction process. The ellipse demarcates 95% confidence interval around group mean. Standard refers to the purification of Rubisco. Numbers represent the publication where the work was described, and corresponding details can be found in Supplementary Table 6
In Figure 7, we observe Rubisco plotting in the upper quadrant, close to the experimental precipitate (number 24). When grouping is visualised according to purification methods (Supplementary Figure 2) no distinct separation is observed. This suggests that introduction of any treatment other than mechanical expression of proteins through juicing appears to strongly influence the amino acid profile. It is unclear whether this proximity to pure Rubisco occurs due the more effective nature of an extraction method or rather the lability of non-Rubisco proteins to the non-physiological conditions of the extraction method. More reports of amino acid profiles of protein concentrates generated using methods such as ultrafiltration, solvent precipitation and centrifugation are required to elucidate purification treatment effect on amino acid profiles. Factor contribution to the dimensions in Figures 6 and 7 is provided in Supplementary Table 7 which shows that Dimension 1 was influenced by essential amino acids such as phenylalanine, valine, histidine, isoleucine and leucine. Dimension 2 was driven primarily by the purity of the protein isolate and its lysine content. This suggests that the nutritional quality of the final LPC is a function of the extraction method employed to obtain it.
Relevance to food production (based on Scottish model)
In Scotland, only \(6.3\text {x}10^{5}\) ha is capable of supporting arable farming, which comprises only about 8% of the total arable land, as shown in Table 6. Although, 70% of the landmass comes under the purview of agriculture, landscape management predominantly pertains to animal husbandry and rough grazing owing to geography [34].
Consequently, the allocation of resources to livestock production is much higher compared to human plant food production as shown in Supplementary Tables 8 and 9. About 1.8 × 10\(^{6}\) ha of land and a plant protein of 9.5 × 10\(^{5}\) t is dedicated to animal husbandry which is about three and eight times greater than food protein requirement respectively. Indigenously produced feed protein can suffice only 24.1% of animal requirement which necessitates supplementation through imports. While the focus of Scottish food production is on premium goods [76, 77] such as high value beef cuts and whisky, the sizeable investments ultimately contributes to only 0.7% to the Scottish GDP. High cost of products restricts access even to local population owing to socio-economic disparity [78].
Furthermore, conversion efficiency of animal sources is shown in Table 7, (detailed in Supplementary Table 10). The average efficiency was found to be 23.7% with monogastrics such as pig, fish and poultry showing higher efficiencies in converting feed to food protein. Milk is the only ruminant product with efficiencies comparable to monogastric products. The total protein used for animal feed is 3.9\(\times 10 ^{6}\) t (including imports). The total protein obtained from terrestrial animal products is about 1.8\(\times 10^{5}\) t, which is about 83% greater than the Scottish population’s requirement.
Despite the meagre investment, the total protein produced is 6.7 × 10\(^{5}\) t, which is 55% greater than the population’s dietary requirement (currently overproducing by 68% when including fisheries). A consequence of poor investment towards cultivation of plant food protein can be observed in Table 8 where a ratio of essential amino acid indigenously produced to that required by the Scottish population is expressed.
Based on the emission data shown in Supplementary Table 11, carbon emissions associated to food production is 8.4 × 10\(^{6}\) t, accounting for 15% of Scotland’s total emission. The average emission associated to animal protein is approximately ten times greater than that of plant protein (102.1 kg CO\(_{2}\) eq/kg animal protein versus 13.4 kg CO\(_{2}\) eq/kg plant protein). Animal husbandry directly contributes to around two-thirds of the carbon emissions associated to food production [79]. In terms of monetary justification, since other economic sectors have a far greater contribution to the GDP, emission per unit currency is shown in Table 9. For every \(\pounds \) earned through agriculture, about 19.7 kg of CO\(_{2}\) eq. emissions are produced. Agriculture is by far the most carbon costly means of Scottish income.
The excessive dependence on animal husbandry as a source of high-quality protein is not only undesirable from an environmental perspective, but also leaves the system vulnerable to shocks such as pandemics [80], trade restrictions and political volatility [81] with trading partners. Much of the protein produced in Scotland is animal-based and even the plant-based protein is primarily sourced from grains such as barley (Supplementary Tables 6 and 7). Furthermore, feed stocks are highly dependent on imports from countries such as Brazil, where soy and high value protein stocks are unsustainably produced through slash-and-burn farming [82–84] resulting in a great loss of local ecological diversity found in the Amazons [85].
With current flux in government policies [86] and lack of economic incentives, sustainable production may be further affected, increasing reliance on imports [87]. Thus, Scottish food production has low economic contribution, a high carbon impact and requires heavy supplementation through imports. In terms of employment, only 0.89% of permanent labour force (around 24,000 persons) and 2.39% including part-time and seasonal labour are directly involved in food production [33]. The status quo necessitates a reflection on the current state of the agricultural setup and work towards raising its economic and ecological sustainability which can most quickly be realised through the adoption of unconventional and diverse means of production.
Conclusions
Through the careful selection of assays amenable to high-throughput routine analysis, it is possible to screen a wide variety of plant species for their potential to contribute to protein provision and the wider cyclic economy. This could greatly enhance screening capabilities with minimal hardware and processing cost. Three invasive plant species, namely, Gorse (Ulex europaeus), Broom (Cystisus scoparius) and Fireweed (Chamaenerion angustifolium), were identified as viable candidates and leaf protein concentrates were produced through extraction and purification technologies. Protein recoveries and purity values suggest Gorse as a suitable candidate for further revalorisation efforts and warrant pilot scale trials to replicate and better account for all associated monetary and carbon costs.
Literature survey of previously published work on LPC methods reveal protein recovery and amino acid profiles to be sensitive to extraction procedures. The efficacy of purification procedures on the other hand was strongly linked to the initial protein content of the plant material. Comparison of amino acid profile reveals that the nutritional quality of final LPC is consequently a function of the extraction method employed to produce it. Characterisation and careful selection and plant material and extraction process is paramount to developing an efficient process for LPC production.
References
B. Bajželj, K.S. Richards, J.M. Allwood, P. Smith, J.S. Dennis, E. Curmi, C.A. Gilligan, Importance of food-demand management for climate mitigation. Nat. Clim. Change 4(10), 924–929 (2014). https://doi.org/10.1038/nclimate2353 (Number: 10)
M. Ercsey-Ravasz, Z. Toroczkai, Z. Lakner, J. Baranyi, Complexity of the international agro-food trade network and its impact on food safety. PLoS ONE 7(5), e37810 (2012). https://doi.org/10.1371/journal.pone.0037810 (Number: 5)
P. (Nel) Wognum, H. Bremmers, J.H. Trienekens, J.G.A.J. van der Vorst, J.M. Bloemhof, Systems for sustainability and transparency of food supply chains: current status and challenges. Adv. Eng. Inf. 25(1), 65–76 (2011). https://doi.org/10.1016/j.aei.2010.06.001
B. Bajželj, K. Richards, The positive feedback loop between the impacts of climate change and agricultural expansion and relocation. Land 3(3), 898–916 (2014). https://doi.org/10.3390/land3030898
P. Gennari, Y. Seid, S. Sorrenti, D.K Navarro, L. De Simone, V. Bizier, Y. Li, A. Charef, M. D’Orazio, R.,Kamana, C.A. Khalil, S. Ditlecadet, S.: FAO-Statistics: Data on Global Agricultural Domains (2019). http://www.fao.org/faostat/en/
A. Iyer, C.S. Bestwick, S.H. Duncan, W.R. Russell, Invasive plants are a valuable alternate protein source and can contribute to meeting climate change targets. Front. Sustain. Food Syst. 5, 575056 (2021). https://doi.org/10.3389/fsufs.2021.575056
C.F. Pratt, K.L. Constantine, S.T. Murphy, Economic impacts of invasive alien species on African smallholder livelihoods. Glob. Food Secur. 14, 31–37 (2017). https://doi.org/10.1016/j.gfs.2017.01.011
BBC: The Visual and Data Journalism Team: The Amazon in Brazil is on fire: how bad is it? https://www.bbc.com/news/world-latin-america-49433767 (2019)
T. Nearmy, Ten Impacts of the Australian Bushfires (2020). https://www.unep.org/news-and-stories/story/ten-impacts-australian-bushfires
C. Gillezeau, M. van Gerwen, R.M. Shaffer, I. Rana, L. Zhang, L. Sheppard, E. Taioli, The evidence of human exposure to glyphosate: a review. Environ. Health 18(1), 2 (2019). https://doi.org/10.1186/s12940-018-0435-5 (Number: 1)
K. Lugowska, The effects of Roundup on gametes and early development of common carp (Cyprinus carpio L.). Fish Physiol. Biochem. 44(4), 1109–1117 (2018). https://doi.org/10.1007/s10695-018-0498-9 (Number: 4)
A. Clemente, M.C. Arques, M. Dalmais, C. Le Signor, C. Chinoy, R. Olias, T. Rayner, P.G. Isaac, D.M. Lawson, A. Bendahmane, C. Domoney, Eliminating anti-nutritional plant food proteins: the case of seed protease inhibitors in pea. PLOS ONE 10(8), e0134634 (2015). https://doi.org/10.1371/journal.pone.0134634
R.C. Merkel, K.R. Pond, J.C. Burns, D.S. Fisher, Intake, digestibility and nitrogen utilization of three tropical tree legumes. Anim. Feed Sci. Technol. 82(1–2), 107–120 (1999). https://doi.org/10.1016/S0377-8401(99)00095-4 (Number: 1–2)
M. Henchion, M. Hayes, A. Mullen, M. Fenelon, B. Tiwari, Future protein supply and demand: strategies and factors influencing a sustainable equilibrium. Foods 6(7), 53 (2017). https://doi.org/10.3390/foods6070053 (Number: 7)
M. Manneberg, H. Lahm, M. Fountoulakis, Oxidation of cysteine and methionine residues during acid hydrolysis of proteins in the presence of sodium azide. Anal. Biochem. 224(1), 122–127 (1995). https://doi.org/10.1006/abio.1995.1016 (Number: 1)
Y.H. Tuan, Optimized determination of cystine/cysteine and acid-stable amino acids from a single hydrolysate of casein- and sorghum-based diet and digesta samples. J. Agric. Food Chem. 45, 3535–3540 (1997)
H..S.. Olcott, H. Frankel-Conrat, Formation and loss of cysteine during acid hydrolysis of protein. Role of tryptophan. J. Biol. Chem. 171, 583–594 (1947) http://www.jbc.org/content/171/2/583.citation
J. Cuq, J. Cheftel, Tryptophan degradation during heat treatments: Part 1—the degradation of free tryptophan. Food Chem. 12(1), 1–14 (1983). https://doi.org/10.1016/0308-8146(83)90045-6 (Number: 1)
J. Cuq, M. Vié, J. Cheftel, Tryptophan degradation during heat treatments: Part 2—Degradation of protein-bound tryptophan. Food Chem. 12(2), 73–88 (1983). https://doi.org/10.1016/0308-8146(83)90019-5 (Number: 2)
H.K. Nielsen, D. De Weck, P.A. Finot, R. Liardon, R.F. Hurrell, Stability of tryptophan during food processing and storage: 1. Comparative losses of tryptophan, lysine and methionine in different model systems. Br. J. Nutr. 53(2), 281–292 (1985). https://doi.org/10.1079/BJN19850035 (Number: 2)
A.G. Dumont, D.S. Boyce, Leaf protein production and use on the farm: an economic study. Grass Forage Sci. 31(4), 153–163 (1976). https://doi.org/10.1111/j.1365-2494.1976.tb01125.x (Number: 4)
N.W. Pirie, The production and use of leaf protein. Proc. Nutr. Soc. 28(1), 85–91 (1969). https://doi.org/10.1079/PNS19690017 (Number: 1)
N.W. Pirie, Leaf protein as human food. Lancet 2(7109), 961–962 (1959). https://doi.org/10.1016/s0140-6736(59)91603-4
S. Nagy, L. Telek, N.T. Hall, R.E. Berry, Potential food uses for protein from tropical and subtropical plant leaves. J. Agric. Food Chem. 26(5), 1016–1028 (1978). https://doi.org/10.1021/jf60219a028 (Number: 5)
L. Telek, Leaf Protein Extraction From Tropical Plants. Plants: The potentials of Extracting Protein, Medicines and Other usaeful Chemicals - Workshop Proceedings pp. 78–108 (1973)
M. Fafunso, M. Byers, Effect of pre-press treatments of vegetation on the quality of the extracted leaf protein. J. Sci. Food Agric. 28(4), 375–380 (1977). https://doi.org/10.1002/jsfa.2740280409 (Number: 4)
N. Broadfield, M.T. McHenry, A world of gorse: persistence of Ulex europaeus in managed landscapes. Plants 8(11), 523 (2019)
Global Invasive Species Database (GISD): Account for Cytisus scoparius (2015). http://www.iucngisd.org/gisd/species.php?sc=441
Global Invasive Species Database (GISD): Account for Ulex europeaus (2015). http://193.206.192.138/gisd/species.php?sc=69
N.C. Ellstrand, S.M. Heredia, J.A. Leak-Garcia, J.M. Heraty, J.C. Burger, L. Yao, S. Nohzadeh-Malakshah, C.E. Ridley, Crops gone wild: evolution of weeds and invasives from domesticated ancestors. Evol. Appl. 3(5–6), 494–504 (2010). https://doi.org/10.1111/j.1752-4571.2010.00140.x (Number: 5–6)
Scottish Government: Scottish agriculture: economic reports (2018). https://www.gov.scot/collections/economic-report-on-scottish-agriculture/
Scottish Government: Scottish Quarterly GDP (2019). https://www.gov.scot/publications/about-gdp/
Scottish Government: Scottish Farm Labour (2016). https://www.webarchive.org.uk/wayback/archive/3000/https://www.gov.scot/Topics/Statistics/Browse/Agriculture-Fisheries/agritopics/farmlabour
The James Hutton Institute: Land Capability for Agriculture in Scotland (2019). https://www.hutton.ac.uk/learning/exploringscotland/land-capability-agriculture-scotland
U.S. Food and Drug: CFR - Code of Federal Regulations Title 21 (2020). https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?fr=172.320
Techniques and Procedures for Collecting, Preserving, Processing, and Storing botanical Specimens. Technical report, Victoria
V.J. Harding, R.M. MacLean, The ninhydrin reaction with amines and amides. J. Biol. Chem. 25, 337–350 (1916)
M.M. Bradford, A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72(1–2), 248–254 (1976). https://doi.org/10.1016/0003-2697(76)90527-3 (Number: 1-2)
P. Smith, R. Krohn, G. Hermanson, A. Mallia, F. Gartner, M. Provenzano, E. Fujimoto, N. Goeke, B. Olson, D. Klenk, Measurement of protein using bicinchoninic acid. Anal. Biochem. 150(1), 76–85 (1985). https://doi.org/10.1016/0003-2697(85)90442-7 (Number: 1)
B.S. Antharavally, K.A. Mallia, P. Rangaraj, P. Haney, P.A. Bell, Quantitation of proteins using a dye-metal-based colorimetric protein assay. Anal. Biochem. 385(2), 342–345 (2009). https://doi.org/10.1016/j.ab.2008.11.024 (Number: 2)
V. Potty, Determination of proteins in the presence of phenols and pectins. Anal. Biochem. 29(3), 535–539 (1969). https://doi.org/10.1016/0003-2697(69)90339-X (Number: 3)
G..E.. Lester, K..S.. Lewers, M..B.. Medina, R..A.. Saftner, Comparative analysis of strawberry total phenolics via Fast Blue BB vs. Folin–Ciocalteu: assay interference by ascorbic acid. J. Food Compos. Anal. 27(1), 102–107 (2012). https://doi.org/10.1016/j.jfca.2012.05.003 (Number: 1)
I.U. Pueyo, M.I. Calvo, Assay conditions and validation of a new UV spectrophotometric method using microplates for the determination of polyphenol content. Fitoterapia 80(8), 465–467 (2009)
M. Lever, Colorimetric and fluorometric carbohydrate determination with p-hydroxybenzoic acid hydrazide. Biochem. Med. 7(2), 274–281 (1973). https://doi.org/10.1016/0006-2944(73)90083-5 (Number: 2)
M. Lever, Carbohydrate determination with 4-hydroxybenzoic acid hydrazide (PAHBAH): effect of bismuth on the reaction. Anal. Biochem. 81(1), 21–27 (1977). https://doi.org/10.1016/0003-2697(77)90594-2
T. Hernandez, A. Hernandez, Available carbohydrates in alfalfa leaf protein concentrates. J. Agric. Food Chem. 42(8), 1747–1749 (1994). https://doi.org/10.1021/jf00044a033 (Number: 8)
P.S. Chow, S.M. Landhausser, A method for routine measurements of total sugar and starch content in woody plant tissues. Tree Physiol. 24(10), 1129–1136 (2004). https://doi.org/10.1093/treephys/24.10.1129 (Number: 10)
M. DuBois, K.A. Gilles, J.K. Hamilton, P.A. Rebers, F. Smith, Colorimetric method for determination of sugars and related substances. Anal. Chem. 28(3), 350–356 (1956). https://doi.org/10.1021/ac60111a017 (Number: 3)
C. Zhang, J.P. Sanders, M.E. Bruins, Critical parameters in cost-effective alkaline extraction for high protein yield from leaves. Biomass Bioenergy 67, 466–472 (2014)
C. Zhang, J.P.M. Sanders, T.T. Xiao, M.E. Bruins, How does alkali aid protein extraction in green tea leaf residue: a basis for integrated biorefinery of leaves. PLOS ONE 10(7), e0133046 (2015). https://doi.org/10.1371/journal.pone.0133046 (Number: 7)
A.G. Calder, K.E. Garden, S.E. Anderson, G.E. Lobley, Quantitation of blood and plasma amino acids using isotope dilution electron impact gas chromatography/mass spectrometry with U-(13)C amino acids as internal standards. Rapid Commun. Mass Spectrom. 13(21), 2080–2083 (1999). https://doi.org/10.1002/(SICI)1097-0231(19991115)13:21<2080::AID-RCM755>3.0.CO;2-O (Number: 21)
H.N. Englyst, J.H. Cummings, Simplified method for the measurement of total non-starch polysaccharides by gas-liquid chromatography of constituent sugars as alditol acetates. Analyst 109(7), 937 (1984). https://doi.org/10.1039/an9840900937 (Number: 7)
H.N. Englyst, G.J. Hudson, The classification and measurement of dietary carbohydrates. Food Chem. 57(1), 15–21 (1996). https://doi.org/10.1016/0308-8146(96)00056-8 (Number: 1)
L.D. Melton, B.G. Smith, Determination of the uronic acid content of plant cell walls using a colorimetric assay. Curr. Protoc. Food Anal. Chem. (2001). https://doi.org/10.1002/0471142913.fae0303s00
R Core Team: R: A Language and Environment for Statistical Computing (2020). https://www.R-project.org/. Programmers: \_:n7004
A. Kassambara, K. Mundt, factoextra: Extract and Visualize the Results of Multivariate Data Analyses (2020). https://CRAN.R-project.org/package=factoextra. Programmers: \_:n7080
H. Wickham, Ggplot2: Elegant Graphics for Data Analysis (Springer, New York, 2016)
T. Wei, V. Simko, R package corrplot: Visualization of a Correlation Matrix (2017). https://github.com/taiyun/corrplot. Programmers: \_:n7073
A. Field, Z. Field, J. Miles, Comparing several means: ANOVA (GLM1), in Discovering Statistics Using R. (Sage, New Delhi, 2012), pp. 398–461
K. Kwon, D. Bae, K. Park, K. Rhee, Aqueous extraction and membrane techniques improve coconut protein concentrate functionality. J. Food Sci. 61(4), 753–756 (1996). https://doi.org/10.1111/j.1365-2621.1996.tb12197.x (Number: 4)
A.T. Culbertson, J.J. Ehrlich, J.Y. Choe, R.B. Honzatko, O.A. Zabotina, Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers. Proc. Natl. Acad. Sci. 115(23), 6064–6069 (2018). https://doi.org/10.1073/pnas.1801105115 (Number: 23)
E. Novaes, M. Kirst, V. Chiang, H. Winter-Sederoff, R. Sederoff, Lignin and biomass: a negative correlation for wood formation and lignin content in trees. Plant Physiol. 154(2), 555–561 (2010). https://doi.org/10.1104/pp.110.161281 (Number: 2)
A.H. Gordon, J.A. Lomax, K. Dalgarno, A. Chesson, Preparation and composition of mesophyll, epidermis and fibre cell walls from leaves of perennial ryegrass (Lolium perenne) and Italian ryegrass (Lolium multiflorum). J. Sci. Food Agric. 36(7), 509–519 (1985). https://doi.org/10.1002/jsfa.2740360702
M. Busse-Wicher, T.C.F. Gomes, T. Tryfona, N. Nikolovski, K. Stott, N.J. Grantham, D.N. Bolam, M.S. Skaf, P. Dupree, The pattern of xylan acetylation suggests xylan may interact with cellulose microfibrils as a twofold helical screw in the secondary plant cell wall of Arabidopsis thaliana. Plant J. 79(3), 492–506 (2014)
Y.W. Sari, W.J. Mulder, J.P.M. Sanders, M.E. Bruins, Towards plant protein refinery: Review on protein extraction using alkali and potential enzymatic assistance. Biotechnol. J. 10(8), 1138–1157 (2015). https://doi.org/10.1002/biot.201400569. (Number: 8)
E.S.M. Abdel-Aal, A.A. Shehata, A.R. El-Mahdy, M.M. Youssef, Extractability and functional properties of some legume proteins isolated by three different methods. J. Sci. Food Agric. 37(6), 553–559 (1986). https://doi.org/10.1002/jsfa.2740370608
A. Moure, J. Sineiro, H. Domnguez, Extraction and functionality of membrane-concentrated protein from defatted Rosa rubiginosa seeds. Food Chem. 74(3), 327–339 (2001). https://doi.org/10.1016/S0308-8146(01)00185-6
R. Pacheco, C. Silva, Global warming potential of biomass-to-ethanol: review and sensitivity analysis through a case study. Energies 12(13), 2535 (2019). https://doi.org/10.3390/en12132535 (Number: 13)
J.C. Claros Garcia, E. Von Sperling, Greenhouse gas emissions from sugar cane ethanol: Estimate considering current different production scenarios in Minas Gerais, Brazil. Renew. Sustain. Energy Rev. 72, 1033–1049 (2017). https://doi.org/10.1016/j.rser.2017.01.046
L. Du, P.J. Arauzo, M.F. Meza Zavala, Z. Cao, M.P. Olszewski, A. Kruse, Towards the properties of different biomass-derived proteins via various extraction methods. Molecules 25(3), 488 (2020). https://doi.org/10.3390/molecules25030488 (Number: 3)
H.P. Makkar, G. Francis, K. Becker, Protein concentrate from Jatropha curcas screw-pressed seed cake and toxic and antinutritional factors in protein concentrate. J. Sci. Food Agric. 88(9), 1542–1548 (2008). https://doi.org/10.1002/jsfa.3248
N. Gerliani, R. Hammami, M. Aïder, Assessment of the extractability of protein-carbohydrate concentrate from soybean meal under acidic and alkaline conditions. Food Biosci. 28, 116–124 (2019). https://doi.org/10.1016/j.fbio.2019.01.004
R.A. Mills, Performance of a forage harvester/dejuicer for leaf protein extraction. Trans. ASAE 27(6), 1634–1637 (1984). https://doi.org/10.13031/2013.33017 (Number 6)
B.E. Knuckles, E.M. Bickoff, G.O. Kohler, PRO-XAN process. Methods for increasing protein recovery from alfalfa. J. Agric. Food Chem. 20(5), 1055–1057 (1972). https://doi.org/10.1021/jf60183a020
R.H. Edwards, R.E. Miller, D. De Fremery, B.E. Knuckles, E.M. Bickoff, G.O. Kohler, Pilot plant production of an edible white fraction leaf protein concentrate from alfalfa. J. Agric. Food Chem. 23(4), 620–626 (1975). https://doi.org/10.1021/jf60200a046 (Number: 4)
F. Akaichi, S. de Grauw, P. Darmon, C. Revoredo-Giha, Does Fair Trade Compete with Carbon Footprint and Organic Attributes in the Eyes of Consumers? Results from a Pilot Study in Scotland, The Netherlands and France. Journal of Agricultural and Environmental Ethics 29(6), 969–984 (2016). https://doi.org/10.1007/s10806-016-9642-7
P. Leat, C. Revoredo-Giha, Risk and resilience in agri-food supply chains: the case of the ASDA PorkLink supply chain in Scotland. Supply Chain Manag. 18(2), 219–231 (2013). https://doi.org/10.1108/13598541311318845 (Number: 2)
C.E. Caspi, G. Sorensen, S. Subramanian, I. Kawachi, The local food environment and diet: A systematic review. Health Place 18(5), 1172–1187 (2012). https://doi.org/10.1016/j.healthplace.2012.05.006
S.H. Duncan, A. Iyer, W.R. Russell, Impact of protein on the composition and metabolism of the human gut microbiota and health. Proc. Nutr. Soc. (2020). https://doi.org/10.1017/S0029665120008022
J.E. Hobbs, Food supply chains during the COVID-19 pandemic. Can. J. Agric. Econ. 68(2), 171–176 (2020). https://doi.org/10.1111/cjag.12237 (Number: 2)
J. Clapp, E. Helleiner, Troubled futures? The global food crisis and the politics of agricultural derivatives regulation. Rev. Int. Polit. Econ. 19(2), 181–207 (2012). https://doi.org/10.1080/09692290.2010.514528 (number 2)
N.C. Brady, Alternatives to slash-and-burn: a global imperative. Agric. Ecosyst. Environ. 58(1), 3–11 (1996). https://doi.org/10.1016/0167-8809(96)00650-0 (Number: 1)
P. Kleinman, The ecological sustainability of slash-and-burn agriculture. Agric. Ecosyst. Environ. 52(2–3), 235–249 (1995). https://doi.org/10.1016/0167-8809(94)00531-I (Number: 2–3)
C. Uhl, Factors controlling succession following slash-and-burn agriculture in Amazonia. J. Ecol. 75(2), 377 (1987). https://doi.org/10.2307/2260425 (Number: 2)
A. Molotoks, E. Stehfest, J. Doelman, F. Albanito, N. Fitton, T.P. Dawson, P. Smith, Global projections of future cropland expansion to 2050 and direct impacts on biodiversity and carbon storage. Glob. Change Biol. 24(12), 5895–5908 (2018)
E. Downing, S. Coe, Brexit: Future UK Agriculture Policy. Briefing Paper CBP 8218, House of Commons, UK (2018). www.parliament.uk/commons-library. Issue: CBP 8218
U.K Government, Import and export UK Agricultural Policy goods (2021). https://www.gov.uk/government/collections/import-and-export-uk-agricultural-policy-goods
Acknowledgements
We would like to thank Phyllis Nicol and Fergus Nicol for their support with protein estimation assays and Teresa Grohmann for helping with manuscript preparation.
Funding
This work was funded by the Scottish Government via RESAS as part of its strategic research programme.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the Supplementary information.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Iyer, A., Guerrier, L., Leveque, S. et al. High throughput method development and optimised production of leaf protein concentrates with potential to support the agri-industry. Food Measure 16, 49–65 (2022). https://doi.org/10.1007/s11694-021-01136-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11694-021-01136-w