Abstract
Background, aim, and scope
Gene expression analyses with real-time (RT)-polymerase chain reaction (PCR) gains importance in marine monitoring. This new technique has to be compared to the classical approaches like the well known biomarker ethoxyresorufin-O-deethylase (EROD) to test their suitability for monitoring programmes. The goal of the present study is to compare EROD activity and CYP1A1 mRNA expression in the important monitoring fish species dab (Limanda limanda) and to answer the question of whether these parameters reflect the polycyclic aromatic hydrocarbon (PAH) contamination of the fish. Further on, glyceraldehyd-3-phosphate dehydrogenase (GAPDH) was investigated as a potential housekeeping gene.
Materials and methods
Female dab were caught in the summer of 2004 in the North Sea and in the Baltic. EROD activity was determined in liver samples by a kinetic fluorimetric assay according to a standard protocol. The gene expression of CYP1A (cytochrome P450 1A) and GAPDH were determined by means of RT-PCR. Results were compared to gonado somatic index and to the concentration of PAH metabolite 1OHPyr (1-hydroxypyrene) analysed in the bile fluids of the fish, respectively.
Results
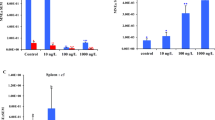
Dab from all stations showed a considerable individual variation in the levels of both CYP1A mRNA and EROD. Highest mean values for CYP1A mRNA and EROD were detected in the northern part of the sampling area. In contrast, the PAH metabolite 1OHPyr was found at the highest concentration in fish caught near the German coast. CYP1A mRNA and EROD showed only a minor but significant correlation (r = 0.32, p < 0.05, n = 123). 1OHPyr in bile correlated significantly (p < 0.05) with the amount of GAPDH mRNA content in the liver.
Discussion
The significant but low correlation of CYP1A mRNA and EROD activity on an individual basis illustrates that these two parameters are apparently not closely linked. However, maximum EROD values correspond with maximum CYP1A mRNA concentrations when station means are regarded. Because EROD and CYP1A mRNA in dab follow different physiological principles, their application will lead to related but not identical monitoring results. This should be taken into account when future marine monitoring programmes are designed. The results also indicate that PAH are not the crucial factor for CYP1A and EROD levels in dab from the off-shore areas in the North Sea. This is remarkable because the PAH metabolism is known to be CYP1A-dependent and the widely used biomarker EROD has been recommended for monitoring PAH-related effects in fish from the North Sea. Due to a correlation between GAPDH and 1OHPyr, GAPDH was not suitable as housekeeping gene for dab.
Conclusions
Neither the results from EROD nor from CYP1A1 mRNA measurements in dab reflected their exposure to PAH as measured by the PAH metabolite 1OHPyr. Thus, the question arises of whether EROD or CYP1A mRNA is a suitable biomarker at all to indicate PAH exposure in dab from the open North Sea.
Recommendations and perspectives
For future biological effect monitoring, it is advisable to measure more and predominately independent parameters by RT-PCR and to incorporate more components of the detoxification system.

Similar content being viewed by others
References
Barber RD, Harmer DW, Coleman RA, Clark BJ (2005) GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues. Physiol Genomics 21:389–395
Biselli S, Reineke N, Heinzel N, Kammann U, Franke S, Hühnerfuss H, Theobald N (2005) Bioassay-directed fractionation of organic extracts of marine surface sediments from the North and Baltic Sea—Part I: Determination and identification of organic pollutants. J Soils Sediment 5(3):171–181
Bowman CJ, Kroll KJ, Hemmer MJ, Folmar LC, Denslow ND (2000) Estrogen-induced vitellogenin mRNA and protein in sheepshead minnow (Cyprinodon variegatus). Gen Comp Endoc 120:300–313
Brack W, Klamer HJC, López de Alda M, Barceló D (2007) Effect directed Analysis of key toxicants in European river basins—a review. Environ Sci Pollut Res 14(1):30–38
Bucheli TD, Fent K (1995) Induction of cytochrome P450 as a biomarker for environmental contamination in aquatic ecosystems. Crit Rev Environ Sci Technol 25:201–261
Craft JA, Robertson FE, McPhail ME, Brown E, Stagg RM (2001) Measurement of cytochrome P4501A induction in dab (Limanda limanda) and other teleosts with species-specific cDNA probes: isolation and characterisation of dab cDNA and its use in expression studies with beta-naphthoflavone-treated fish. Comp Biochem Phys C 129(2):115–27
Fernandes JM, Mommens M, Hagen O, Babiak I, Solberg C (2008) Selection of suitable reference genes for real-time PCR studies of Atlantic halibut development. Comp Biochem Physiol B Biochem Mol Biol 150:23–32
Folmar LC, Hemmer M, Hemmer R, Bowman C, Kroll K, Denslow ND (2000) Comparative estrogenicity of estradiol, ethinyl estradiol and diethylstilbestrol in vivo, male sheepshead minnow (Cyprinodon variegatus), vitellogenin bioassay. Aquat Toxicol 49:77–88
George S, Gubbins M, MacIntosh A, Reynolds W, Sabine V, Scott A, Thain J (2004) A comparison of pollutant biomarker responses with transcriptional responses in European flounders (Platichthys flesus) subjected to estuarine pollution. Mar Environ Res 58:571–575
Griffin TJ, Gygi SP, Ideker T, Rist B, Eng J, Hood L, Aebersold R (2002) Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics 1:323–333
Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R (2001) Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292:929–934
Kammann U (2007) PAH metabolites in bile fluids of dab (Limanda limanda) and flounder (Platichthys flesus)—spatial distribution and seasonal changes. Environ Sci Pollut Res 14(2):102–108
Kammann U, Lang T, Vobach M, Wosniok W (2005) Ethoxyresorufin-O-deethylase (EROD) activity in dab (Limanda limanda) as biomarker for marine monitoring. Environ Sci Pollut Res 12(3):140–145
Lange U, Saborowski R, Siebers D, Buchholz F, Karbe L (1998) Temperature as a key factor determining the regional variability of the xenobiotic-inducible ethoxyresorufin-O-deethylase activity in the liver of dab (Limanda limanda). Can J Fish Aquat Sci 55(2):328–338
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with a Folin phenol reagent. J Biol Chem 193:265–275
Marchand J, Tanguy A, Charrier G, Quiniou L, Plee-Gauthier E, Laroche J (2006) Molecular identification and expression of differentially regulated genes in the European flounder, Platichthys flesus, submitted to pesticide exposure. Mar Biotechnol 8:275–294
OSPAR (1997) JAMP guidelines for monitoring contaminants in biota. OSPAR Oslo and Paris Commission, London
OSPAR (1998a) JAMP guidelines for general biological effects monitoring. OSPAR Oslo and Paris Commission, London
OSPAR (1998b) JAMP guidelines for contaminant-specific biological effects monitoring. OSPAR Oslo and Paris Commission, London
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45
Quiros L, Pina B, Sole M, Blasco J, Loper MA, Riva MC, Barcelo D, Raldua D (2007) Environmental monitoring by gene expression biomarkers in Barbus graellsii: Laboratory and field studies. Chemosphere 67:1144–1154
Roberts A, Oris JT, Burton Jr GA, Clements WH (2005) Gene expression in caged fish as a first-tier indicator of contaminant exposure in streams. Environ Toxicol Chem 24(12):3092–3098
Sancho E, Ferrando MD, Fernandez C, Andreu E (1998) Liver energy metabolism of Anguilla anguilla after exposure to fenitrothion. Ecotoxicol Environ Saf 41:168–175
Sirover MA (2005) New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J Cell Biochem 95(1):45–52
Stagg RM, McIntosh A (1998) Biological effects of contaminants: Determination of CYP1A-dependent mono-oxygenase activity in dab by fluorimetric measurement of EROD activity. ICES Techniques in Marine Environmental Sciences, No. 23. International Council for the Exploration of the Sea, Copenhagen
Tang R, Dodd A, Lai D, McNabb WC, Love DR (2007) Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim Biophys Sin 39:384–390
Timme-Laragy AR, Cockman CJ, Matson CW, Di Giulio RT (2007) Synergistic induction of AHR regulated genes in developmental toxicity from co-exposure to two model PAHs in zebrafish. Aquat Toxicol 85:241–250
Tom M, Auslander M (2005) Transcript and protein environmental biomarkers in fish—a review. Chemosphere 59:155–162
Tom M, Shmul M, Shefer E, Chen N, Slor H, Rinkevich B, Herut B (2003) Quantitative evaluation of hepatic cytochrome P4501A transcript, protein, and catalytic activity in the striped sea bream (Lithognathus mormyrus). Environ Toxicol Chem 22(9):2088–2092
Van der Oost R, Beyer J, Vermeulen NPE (2003) Fish bioaccumulation and biomarkers in environmental risk assessment: a review. Environ Toxicol Phar 13(2):57–149
Whyte JJ, Jung RE Schmitt CJ, Tillitt DE (2000) Ethoxyresorufin-O-deethylase (EROD) activity in fish as a biomarker of chemical exposure. Crit Rev Toxicol 30:347–570
Williams TD, Diab AM, George SG, Sabine V, Chipman JK (2007) Gene expression responses of European flounder (Platichthys flesus) to 17-β estradiol. Toxicol Lett 168:236–248
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Magnus Engwall
Rights and permissions
About this article
Cite this article
Kammann, U., Lang, T., Berkau, AJ. et al. Biological effect monitoring in dab (Limanda limanda) using gene transcript of CYP1A1 or EROD—a comparison. Environ Sci Pollut Res 15, 600–605 (2008). https://doi.org/10.1007/s11356-008-0048-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-008-0048-6