Abstract
Purpose
Estrogen receptors (ER) are implicated in psychiatric disorders. We assessed if ER availability in the human brain could be quantified using 16α-[18F]-fluoro-17β-estradiol ([18F]FES) positron emission tomography (PET).
Procedures
Seven post‑menopausal women underwent a dynamic [18F]FES PET scan with arterial blood sampling. A T1-weighted MRI was acquired for anatomical information. After one week, four subjects received a selective ER degrader (SERD), four hours before the PET scan. Pharmacokinetic analysis was performed using a metabolite-corrected plasma curve as the input function. The optimal kinetic model was selected based on the Akaike information criterion and standard error of estimated parameters. Accuracy of Logan graphical analysis and standardized uptake value (SUV) was determined via correlational analyses.
Results
The reversible two-tissue compartment model (2T4k) model with fixed K1/k2 was preferred. The total volume of distribution (VT) could be more reliably estimated than the binding potential (BPND). A high correlation of VT with Logan graphical analysis was observed, but only a moderate correlation with SUV. SERD administration resulted in a reduced VT in the pituitary gland, but not in other regions.
Conclusions
The optimal quantification method for [18F]FES was the 2T4k with fixed K1/k2 or Logan graphical analysis, but specific binding was only observed in the pituitary gland.
Similar content being viewed by others
Introduction
Estrogen receptors (ER) are associated with regulatory functions in the central nervous system (CNS) and exert neuroprotective and neurotrophic effects [1]. Substantial evidence suggests that estrogens play an important role in psychiatric disorders, such as postmenopausal and postnatal depression [2]. Hormone replacement therapy can improve such postmenopausal complaints [3]. Although the ER is supposedly involved, there is not much known about its exact role in these psychiatric disorders. Estrogen receptor density in the living human brain may be assessed by non-invasive imaging techniques, such as positron emission tomography (PET). Several PET tracers have been developed for the purpose of ER imaging [4]. Among these, 16α-[18F]-fluoro-17β-estradiol ([18F]FES) is the best-characterized tracer with high affinity and selectivity for ER. [18F]FES PET is now used in clinical trials and regular patient care for imaging of ER expression in hormone-sensitive tumors, mainly breast cancer [5, 6], but may also be suitable for ER imaging in the brain. The first study using [18F]FES PET to assess ER expression in rat brains found the highest [18F]FES uptake in pituitary gland and hypothalamus, regions with high ER expression. Co-administration of 17β-estradiol with the tracer led to a decrease in tracer uptake in those two brain regions [7], suggesting that tracer uptake was ER-mediated. Despite these promising preclinical results, [18F]FES PET has not been fully evaluated for imaging of ER expression in the human brain yet. Whole-body PET images of cancer patients show heterogenous uptake (SUV) of the tracer in the brain, with higher uptake in white than grey matter [8, 9], but it is unclear whether this uptake is ER-mediated. The purpose of the current study was to determine if [18F]FES PET can be used for imaging of ER expression in the human brain and to assess what the optimal method is to quantify [18F]FES binding. The selective ER degrader elacestrant was administered to assess whether [18F]FES uptake in the brain is ER-mediated.
Materials and Methods
Experimental Design and Study Set-up
PET imaging was carried out as part of a phase 1 study of the experimental drug elacestrant, which aimed to determine ER availability [10]. Elacestrant was developed as a selective estrogen receptor degrader that crosses the blood–brain barrier for the treatment of estrogen receptor positive breast cancer brain metastases. The drug was shown to competitively bind to ERs [11] in the same binding pocket as estrogen and [18F]FES [12]. The study was approved the independent ethics committee of the foundation “evaluation of ethics in biomedical research” (CCMO code: NL49312.056.14) and was performed in accordance with standards for Good Clinical Practice, in full compliance with the principles of the 1964 Declaration of Helsinki. Seven healthy post-menopausal women (age 61.5 ± 9.7) were included in the study. Informed consent was obtained from all individual participants included in the study. Exclusion criteria were the use of any concomitant medication, smoking or any other substance dependence. At baseline, a 3D T1-weighted MRI and a dynamic [18F]FES PET scan were acquired. Subjects were treated daily with an oral dose of elacestrant (500 mg) for 7 days, to reach steady-state levels in plasma, without hormonal replacement therapy. In 4 subjects, [18F]FES PET was repeated 4 h after the last drug dose.
MRI Acquisition
A structural 3D T1-weighted MRI sequence (matrix size 256 × 256 × 3, voxel size 0.97 × 0.97 × 20, repetition time 11.12 ms, echo time 4.60 ms) on a 3 Tesla Ingenuity TF system (Philips, Netherlands) was acquired for each subject to be used as individual anatomical reference for spatial normalization and co-registration of the PET scans.
PET Acquisition
A catheter was placed in a brachial vein for intravenous administration of the tracer, and a cannula was inserted into the radial artery of the opposite wrist for blood sampling. PET/CT images were acquired with a Biograph mCT system (Siemens, Knoxville, USA). After a low-dose CT was acquired, a bolus (8.3 ml) of [18F]FES (baseline 199 ± 6 MBq; post-dose 209.0 ± 13 MBq) was intravenously injected (0.5 ml/s) and a 90-min dynamic PET scan of the brain was started. The dynamic PET data were reconstructed using a time-of-flight version of the 3D ordered-subsets-expectation–maximization algorithm (3 iterations, 24 subsets) and corrected for decay, attenuation scatter and random coincidences. List-mode data were reconstructed into 33 temporal frames: 6 × 5 s; 4 × 10 s; 4 × 15 s; 3 × 30 s; 3 × 60 s; 4 × 150 s; 3 × 300 s; 6 × 600 s. The final images had a matrix size of 400 × 400 × 111 and a voxel size of 2.03 × 2.03 × 2 mm.
Blood Sampling and Processing
The radioactivity concentration in arterial blood was continuously measured during the first 30 min of the scan, using an automatic blood sampling system (Veenstra Instruments, Joure, Netherlands). In addition, seven manual samples were taken at approximately 5, 10, 20, 30, 40, 60 and 90 min after tracer injection for calibration of the automated sampler and metabolite analysis. In each sample, the radioactivity concentration in 250 µl of whole-blood and plasma were measured with an automated gamma-counter (Wizard2480, PerkinElmer, USA). The radioactivity concentration was expressed as standardized uptake values (SUV). SUV values were calculated by dividing the measured radioactivity concentration (kBq/mL) by the ratio of the injected dose (kBq) and body weight (g) of the subject. It was assumed that 1 g equals 1 mL.
For metabolite analysis, 50 μl aliquots of the plasma samples were diluted with 100 μl of acetonitrile and centrifuged (5 min, 15000 g). A 2.5 µl aliquot of the supernatant, was analyzed by thin-layer chromatography, using a silica gel 60 F254 TLC plate (Merck, Germany) and n-hexane/ethyl acetate (7/3) as the mobile phase. A phosphor storage screen (PerkinElmer, USA) was exposed to the TLC plate for approximately 18 h. The phosphor storage screen was scanned with a Cyclone Imaging System (PerkinElmer, USA) and analyzed with OptiQuant Software version 3.0 to determine the percentage of intact [18F]FES in plasma. A one-exponential function was fitted to the metabolite data and was used to generate a metabolite-corrected plasma input function.
Data Analysis and Image Processing
PET images were co-registered to the corresponding T1-weighted MRI scan of the same subject, using PMOD version 4.1 (PMOD Technologies LLC, Zürich, Switzerland). Head motion correction was applied to the PET data, if necessary. MRI scans were spatially normalized to Montreal Neurological institute (MNI) space [13] and the transformation matrix was used to align the co-registered PET images. Volumes-of-interest (VOIs) for individual brain regions were obtained from the Hammers atlas [14]. The 83 available brain structures within the Hammers atlas were aggregated into 15 brain regions, as no differences between left and right were expected and no differences within cortical regions. Regions with expected high ER expression, e.g., thalamus, hippocampus and amygdala, were analyzed as separate structures. VOIs for white matter, grey matter, and the whole brain were segmented from the MRI dataset in SPM12 (Wellcome Trust Center for Neuroimaging, UK) with a probability map threshold of 0.5. A VOI for the pituitary gland was drawn manually for each subject in PET-space, using a 3D iso-contour at 40% of the maximum uptake. TACs for each VOI were generated for kinetic modeling.
Kinetic Modeling
Pharmacokinetic modeling was performed using the metabolite-corrected plasma TAC as the input function and the TAC of whole-blood for blood volume correction. To improve the accuracy of the fits, frame duration and frame mid-time decay were applied as weighting factors for all evaluations. The one-tissue compartment model (1T2k), irreversible two-tissue compartment model (2T3k) and reversible two-tissue compartment model (2T4k) were assessed for fitting the regional TACs, using a fitted fractional volume of blood (VB). The 2T4k model was further explored using the VB as a fixed value of 0.05 [15] and by fixation of the influx-efflux rate constant ratio (K1/k2) to the K1/k2 ratio of the whole-brain. In addition, Logan graphical analysis was performed with a starting time (t*) of 20 min.
The Akaike Information Criterion (AIC) was used to select the most appropriate model. The standard error in the parameter estimated by the compartment models was used to determine the reliability of in the total volume of distribution (VT) and non-displaceable binding potential (BPND) estimates, using an arbitrary cut-off value of 25%. The accuracy of SUV and Logan graphical analysis derived VT was determined via correlational analyses with the macro-parameters of the optimal compartment model. The change in ER availability by the drug was calculated using the Lassen plot [16].
Statistical Analysis
All statistical analyses were performed using IBM SPSS Statistics (Version 23, Armonk, NY, USA). Differences in kinetic parameter estimates between the baseline and post-dose scans were assessed by generalized estimated equation, using the main effects “brain region” and “treatment” and the interaction “brain region × treatment”. A Bonferroni post-hoc analysis was used for multiple comparisons correction. Results are reported as mean ± standard deviation (SD) and were considered statistically significant if the null hypothesis was rejected at a probability of 95% (p < 0.05). The correlation between parameters from different models were examined by Pearson linear regression analysis.
Results
Tracer Kinetics and Metabolism
Highest [18F]FES concentration in blood and plasma was reached after approximately 2 min, which was followed by a rapid decrease (Fig. 1A). Plasma radioactivity comprised 81 ± 7% and 14 ± 3% of intact tracer at 5 and 90 min, respectively (Fig. 1B). No significant effects of drug administration were observed.
[18F]FES uptake was relatively homogeneously distributed across different brain regions, although uptake was higher in white matter than in grey matter. Highest brain uptake (SUV 3.47 ± 0.37) was observed at 1.3 ± 0.4 min after tracer administration (Fig. 1C). Peak uptake in the pituitary gland was 4.82 ± 0.61. After the initial peak, the tracer was gradually cleared from the brain, resulting in a SUV of 0.40 ± 0.04 in the whole-brain and 1.17 ± 0.24 in pituitary gland at 80–90 min after tracer injection.
Compartmental Models
The 1T2k model did not fit the data properly (data not shown). At baseline, the 2T4k model fitted the regional [18F]FES TACs better than the 2T3k model for the majority of subjects (Fig. 2 and Supplementary Fig. 1). The 2T4k model could not always estimate the outcome parameters with high precision, as 9.0% of VT and 31.6% of BPND values were estimated with a standard error > 25% (Table 1). To increase the precision and robustness of the estimated parameters, the K1/k2 of individual brain regions was fixed to the K1/k2 value of the whole brain and the VB was fixed to 0.05. The K1/k2 values (Supplementary Table 1) ranged from 0.27 to 1.14 for individual subjects (variance of 0.055) but showed less variance over different brain regions (variance of 0.005 to 0.037 (median 0.010) for individual subjects). Although the pituitary is outside the blood–brain barrier, there were no differences between the K1 (p = 0.43), k2 (p = 0.35) and K1/k2 (p = 0.86) values for the whole brain and pituitary and therefore the whole brain K1/k2 was also used for this brain region. The fixation of the K1/k2 ratio resulted in lower AIC values, whereas fixation of VB hardly had any effect. Fixation of the K1/k2 ratio resulted in a reduction of the number of VT estimates at baseline with a standard error > 25% from 9.0% (2T4k) and 6.7% (2T4k-VB) to 7.5% (2T4k-K1k2) and 2.3% (2T4k-K1k2-VB) (Table 1). The fraction of BPND estimates with a standard error > 25% was reduced from 31.6% (2T4k) and 30.8% (2T4k-VB) to 14.3% (2T4k-K1k2), and 19.5% (2T4k-K1k2-VB).
Pearson correlations of the models with constrained parameters with the prime compartmental model (2T4k) were performed to assess the bias introduced by fixation of K1/k2 and VB (Fig. 3). The VT values estimated by the 2T4k model correlated well with the VT estimates from the 2T4k-VB (R2 = 0.99, p < 0.0001), 2T4k-K1k2 (R2 = 0.97, p < 0.0001) and 2T4k-K1k2-VB model (R2 = 0.98, p < 0.0001). Worse correlations (R2) were observed when BPND estimates from the 2T4k model were correlated with BPND estimates derived from the 2T4k-VB (R2 = 0.98, p < 0.0001), 2T4k-K1k2 (R2 = 0.81, p < 0.0001) and 2T4k-K1k2-VB model (R2 = 0.72, p < 0.0001), even if only values with a standard error < 25% were included.
Correlations between VT (A) and BPND (B) values estimated with the 2T4k-VB, 2T4k-K1k2, and 2T4k-K1k2-VB models with the estimates from the 2T4k model. Only VT and BPND estimates with a standard error < 25% were included in the correlations. Highest values in the plots represent measurements for pituitary gland
Considering the AIC, standard error of the estimated parameters, bias and correlation of outcome parameters with cardinal compartmental model, the 2T4k-K1k2 model seems to be the preferred model with VT as the preferred outcome parameter.
Logan Graphical Analysis
Logan graphical analysis could fit the data well. All VT values estimated with Logan graphical analysis had a standard error < 25%. Baseline VT values derived from Logan graphical analysis were strongly correlated with VT values estimated from the 2T4k compartment model (R2 = 0.99, p < 0.0001; Fig. 4) and the 2T4k-K1k2 model (R2 = 0.98, p < 0.0001; Supplementary Fig. 2A), although some underestimation was observed.
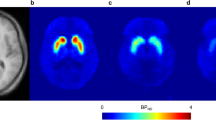
Representative baseline and post-dose VT images derived from Logan graphical analysis are shown in Fig. 5.
Standardized Uptake Value
SUV80-90 values were only moderately correlated with VT values derived for the 2T4k (R2 = 0.61, p < 0.0001; Supplementary Fig. 3) or 2T4k-K1k2 model (R2 = 0.58, p < 0.0001; Supplementary Fig. 2B). Surprisingly, SUV80-90 measurements correlated better with BPND values estimated from the 2T4k-K1k2 model (R2 = 0.90, p < 0.0001; Supplementary Fig. 2C).
Effect of the Drug
The administration of a nonsteroidal selective ER degrader (elacestrant) before the post-dose [18F]FES PET scan reduced the preference for the 2T4k model. The number of subjects and the percentage of brain regions, in which the 2T3k model was preferred based on the AIC values, was similar to those in which the 2T4k model was preferred (Supplementary Fig. 1). Imposing constraints on VB did not substantially improve the robustness of the post-dose outcome parameters (Table 1). However, fixation of the K1/k2 ratio reduced the percentage of estimated VT values with a standard error > 25% from 32.9% (2T4k) and 32.9%, (2T4k-VB) to 14.5% (2T4k-K1k2) and 11.8% (2T4k-K1k2-VB). The percentage of estimated BPND values from post-dose scans with a standard error > 25% was reduced from 89.5% and 85.5% to 32.3% and 36.9%, respectively. The VT values derived from the post-dose scans using the 2T4k-K1k2 model were positively correlated with the VT values assessed with the 2T4k compartmental model (R2 = 0.74, p < 0.0001). In contrast, the BPND values estimated with these models did not show any correlation. Therefore, the 2T4k-K1k2 model with the volume of blood (VB) as a fit parameter was considered the optimal model with the VT as the optimal outcome parameter for both the post-dose and the baseline scans.
The drug administration only resulted in a statistically significant reduction of [18F]FES uptake in pituitary gland, irrespective whether the VT values derived from the 2T4k-K1k2 model or Logan analysis, the BPND from the 2T4k-K1k2 model, or the SUV80-90 were used as outcome parameter (Table 2). Similar results were found when only the four subjects that completed both PET scans were included in the statistical analysis (data not shown). Based on the VT data these subjects, a daily oral dose (500 mg) of elacestrant for 7 days would result in a 64% (VT from 2T4k-K1k2) or 62% (VT from Logan analysis) reduction in ER availability in the pituitary gland 4 h after the last dose.
Discussion
The primary objectives of this study were to investigate the feasibility of using [18F]FES PET for imaging of ER in the human brain, to determine an optimal quantitation approach and to assess the reduction of ER availability after administration of an experimental drug. The 2T4k model was favored over the 1T2k and 2T3k model based on visual assessment of the fit, AIC values and the standard error of the estimated outcome parameters. The 2T4k model provides more robust estimations of the VT than the BPND and therefore VT is the preferred outcome parameter.
Even with the preferred 2T4k model for quantitation of [18F]FES PET, relatively high standard errors, in particular in the estimated BPND, were observed. Fixing the ratio of K1/K2 enhanced the quality of the fits and decreased the frequency of BPND estimates with a standard error > 25% by approximately two-fold. Although fixation of the K1/K2 ratio hardly had any effect on the baseline VT estimates, it did improve the VT estimates from the post-dose scans. The good correlations between the parameter estimated with the 2T4k-K1k2 model and the 2T4k model and the slope of these correlations being close to one imply that fixing of the K1/k2 ratio did not introduce any significant bias in the data analysis. Imposing an additional constraint by fixating the VB to 0.05 increased rather than reduced the standard error in the outcome estimates. A plausible explanation could be that the regional blood flow and blood volume differ between various regions of the human brain [15, 17]. Some brain regions, such as cerebellum, have a blood volume that is smaller than 5%, while regions like frontal-lobe may have a higher blood volume fraction.
Logan graphical analysis could fit data well, confirming the findings from compartment modeling that [18F]FES acts as a reversible PET tracer in the brain. The strong correlation of VT values estimated with Logan graphical analysis with VT values derived from the 2T4k and 2T4k-K1k2 model indicate that Logan graphical analysis could be a good alternative for compartment modeling, as it can more reliably estimate VT values. Yet, Logan graphical analysis may introduce some bias due to underestimation of VT or increase of the noise, as fit parameters would be acquired through nonlinear estimation [18]. The SUV80-90 only moderately correlated with VT values derived from the compartmental model and therefore is not a good measure for [18F]FES binding in the brain.
[18F]FES displayed relatively high uptake in white matter, whereas uptake was relatively low and homogeneously distributed in grey matter regions. The relatively high uptake observed in white matter was not reduced after the administration of the drug, indicating that the uptake in white matter is mainly due to non-specific binding. This can be explained by the relatively high lipophilicity of the PET tracer. Low estrogen receptor expression in white matter was found in autoradiography studies in the female rat and monkey, supporting the lack of specific binding [19, 20]. Administration of elacestrant did not have any effect on the blood kinetics of the tracer or its metabolism but reduced the preference for the 2T4k model in post-dose scans, resulting in an approximately equal number of brain regions in which the 2T3k model was preferred. One reason for this shift in model preference could be that the post-dose scans are less influenced by the second compartment, representing specific binding, due to saturation of the receptor. While in case of complete receptor saturation the 1T2k model would be expected to give a better fit, we did not find this. Apparently, there is still binding of [18F]FES so that the fit is better when the rate constants k3 and k4 are estimated as well. As the signal to noise ratio of PET data in areas with low receptor availability is intrinsically low, it is more difficult to accurately estimate the rate constants, in particular k3 and k4, which could result in the preference for 2T3k.
We could only demonstrate specific binding in pituitary gland, which is in line with findings from animal studies [7, 21]. Administration of elacestrant led to a reduction of ER availability of 62–64% in the pituitary gland. Besides pituitary gland, no statistically significant effect of the drug on [18F]FES binding could be observed in any brain region. This could be due to the relatively low ER density the brain. ER-mediated binding may thus have been obscured by high levels of non-specific binding. Previous studies in rats reported specific [18F]FES binding in hypothalamus [7]. Since the hypothalamus in rats is located close to the pituitary gland, this apparent specific binding might be the result of spillover from the pituitary gland. Another reason why specific binding was only observed in the pituitary gland could be that the pituitary gland is located outside the blood–brain barrier (BBB). [18F]FES or elacestrant might be a substrate for an efflux pump in the BBB and be extruded from the brain before it can reach cerebral ER. Based on the TACs of [18F]FES uptake in the brain, however, it appears that BBB penetration is not impaired, since peak uptake in the whole brain is in the same range as peak uptake in the pituitary gland (SUV 3.5 vs. 4.8). In an intracranial mouse model elacestrant levels in the tumor were comparable to levels observed in plasma, suggesting that elacestrant can cross the BBB [11]. Furthermore, in a clinical study elacestrant was detected in cerebrospinal fluid after oral administration [10]. Finally, it has to be mentioned that the low number of subjects is a limitation of the study. Especially the lack of significant specific binding in other regions than the pituitary gland could be due to the small number of subjects that underwent both the baseline and post-dose scan, which limits the statistical power of the study.
Due to the poor sensitivity to detect specific binding in the brain, [18F]FES does not seem to be a suitable radioligand for measuring the cerebral ER expression in patients with psychiatric or neurological diseases. However, [18F]FES PET could be useful for investigation of the role of ER in the pituitary gland in stress-related disorders. Stress activates the hypothalamic–pituitary–adrenal (HPA) axis. Several studies have shown that the response of the HPA-axis to stress can be modulated by ER-mediated signaling and that fluctuations in circulating estrogen levels affect the activity of the HPA-axis [22, 23]. The role of ER in the pituitary gland largely remains to be elucidated and [18F]FES PET could provide a useful tool for this purpose.
Conclusion
Our study indicates that the reversible 2T4k-K1k2 model is the model of choice to describe the pharmacokinetics of [18F]FES in human brain. Besides the compartment model, Logan graphical analysis can also be applied as a robust approach for quantification of [18F]FES uptake in the human brain. [18F]FES showed ER-mediated uptake in pituitary, but not in the brain. Therefore, [18F]FES only seems to be a suitable PET tracer for quantification of ER expression in tissues with high ER density like pituitary gland.
References
Baulieu E (1998) Neurosteroids: a novel function of the brain. Psychoneuroendocrinology 23:963–987
Hwang WJ, Lee TY, Kim NS, Kwon JS (2020) The role of estrogen receptors and their signaling across psychiatric disorders. Int J Mol Sci 22:373
McCarrey AC, Resnick SM (2015) Postmenopausal hormone therapy and cognition. Horm Behav 74:167–172
Moraga-Amaro R, Doorduin J, Dierckx RA, de Vries EF (2021) PET and SPECT imaging of steroid hormone receptors in the brain. In: Dierckx RA, Otte A, de Vries EF, van Waarde A, Lammertsma AA (eds) PET and SPECT of neurobiological systems. Springer, Cham, pp 483–520
van Kruchten M, Glaudemans AW, de Vries EF et al (2012) PET imaging of estrogen receptors as a diagnostic tool for breast cancer patients presenting with a clinical dilemma. J Nucl Med 53:182–190
Chae SY, Ahn SH, Kim S-B et al (2019) Diagnostic accuracy and safety of 16α-[18F] fluoro-17β-oestradiol PET-CT for the assessment of oestrogen receptor status in recurrent or metastatic lesions in patients with breast cancer: a prospective cohort study. Lancet Oncol 20:546–555
Khayum MA, de Vries EF, Glaudemans AW et al (2014) In vivo imaging of brain estrogen receptors in rats: a 16α-18F-fluoro-17β-estradiol PET study. J Nucl Med 55:481–487
Iqbal R, Yaqub M, Oprea-Lager DE et al (2022) Biodistribution of 18F-FES in patients with metastatic ER+ breast cancer undergoing treatment with Rintodestrant (G1T48), a novel selective estrogen receptor degrader. J Nucl Med 63:694–699
Mankoff DA, Peterson LM, Tewson TJ et al (2001) [18F] fluoroestradiol radiation dosimetry in human PET studies. J Nucl Med 42:679–684
Conlan MG, de Vries EF, Glaudemans A et al (2020) Pharmacokinetic and pharmacodynamic studies of elacestrant, a novel oral selective estrogen receptor degrader, in healthy post-menopausal women. Eur J Drug Metab Pharmacokinet 45:675–689
Garner F, Shomali M, Paquin D et al (2015) RAD1901: a novel, orally bioavailable selective estrogen receptor degrader that demonstrates antitumor activity in breast cancer xenograft models. Anticancer Drugs 26:948–956
Hancock GR, Young KS, Hosfield DJ et al (2022) Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells. NPJ Breast Cancer 8:130–156
Ashburner J, Friston KJ (2005) Unified segmentation. Neuroimage 26:839–851
Hammers A, Allom R, Koepp MJ et al (2003) Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp 19:224–247
Leenders K, Perani D, Lammertsma A et al (1990) Cerebral blood flow, blood volume and oxygen utilization: normal values and effect of age. Brain 113:27–47
Cunningham VJ, Rabiner EA, Slifstein M et al (2010) Measuring drug occupancy in the absence of a reference region: the Lassen plot re-visited. J Cereb Blood Flow Metab 30:46–50
Rostrup E, Knudsen GM, Law I et al (2005) The relationship between cerebral blood flow and volume in humans. Neuroimage 24:1–11
Logan J (2000) Graphical analysis of PET data applied to reversible and irreversible tracers. Nucl Med Biol 27:661–670
Pfaff D, Keiner M (1973) Atlas of estradiol-concentrating cells in the central nervous system of the female rat. J Comp Neurol 151:121–158
Pfaff DW, Gerlach JL, McEwen BS et al (1976) (1976) Autoradiographic localization of hormone-concentrating cells in the brain of the female rhesus monkey. J Comp Neurol 170:279–293
Paquette M, Phoenix S, Lavallée É et al (2020) Cross-species physiological assessment of brain estrogen receptor expression using 18 F-FES and 18 F-4FMFES PET imaging. Mol Imaging Biol 22:1403–1413
Handa RJ, Mani SK, Uht RM (2012) Estrogen receptors and the regulation of neural stress responses. Neuroendocrinology 96:111–118
Heck AL, Handa RJ (2019) Sex differences in the hypothalamic–pituitary–adrenal axis’ response to stress: an important role for gonadal hormones. Neuropsychopharmacology 44:45–58
Acknowledgements
The sponsor had no role in gathering, analyzing, or interpreting the data. The authors thank all the subjects who participated in this study.
Author information
Authors and Affiliations
Contributions
JD, AG, RD and EdV contributed to the design of the work; NG and CvdW analyzed the data; NG, JD, AW, AvW and EdV interpreted the data. NG wrote the manuscript. All authors revised the manuscript, approved the final version and agreed to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standard.
Conflict of Interest
This study was financially supported by Radius Health, Inc. Institutional financial support for contracted research beyond the scope of this work was obtained from GE Healthcare, Hoffmann-La Roche, GlaxoSmithKline, Ionis Pharmaceuticals, Janssen-Cilag, and Bristol Myers Squibb. No other potential conflicts of interest relevant to this article exist.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ghazanfari, N., Doorduin, J., van der Weijden, C.W.J. et al. Pharmacokinetic Analysis of [18F]FES PET in the Human Brain and Pituitary Gland. Mol Imaging Biol 26, 351–359 (2024). https://doi.org/10.1007/s11307-023-01880-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11307-023-01880-z