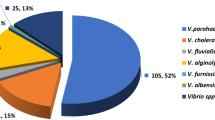
Abstract
Vibrio parahaemolyticus is one of the leading causes of diarrhoea and gastroenteritis in human on consumption of raw or insufficiently cooked seafood. This study was aimed at isolating and characterizing the pathogenic and pandemic V. parahaemolyticus from oysters (n = 90) in coastal parts of West Bengal, India; their antibiotic resistance and potential for involvement in the food chain. During bacteriological culture, typical V. parahaemolyticus colony was recovered in 88.9% samples followed by presumptive identification in 71 (78.9%) samples by characteristic biochemical (K/A) test. All the presumptive isolates (n = 71) were confirmed by species specific Vp-toxR PCR assay. Of these, 10 (14.08%) were tdh+ and none for the trh. Further, 5 (50%) of these tdh+ isolates were found to carry the pandemic potential gene in PGS-PCR assay; however, none in GS-PCR. Majority (80%) of these pathogenic (tdh+) isolates belonged to pandemic serovars (OUT: KUT; OUT: K24; O1: KUT; O1:K25; O10: KUT) and only 20% to non-pandemic serovars (OUT: K15; O9:K17). All the isolates (100%) exhibited resistance to cefpodoxime followed by ampicillin and cefotaxime (90%), ceftizoxime (60%), tetracycline (50%), ceftriaxone (40%), ciprofloxacin and nalidixic acid (10% each). Overall, the study findings suggested that 11.1% (10/90) of commonly marketed oysters in this area were harbouring pathogenic V. parahaemolyticus. Moreover, 5.5% (5/90) of the oyster population were harbouring pandemic strains of this pathogen. Besides, the pathogenic isolates from oysters were exhibiting a considerable genetic relatedness (53 to 70%) to human clinical isolates in PFGE analysis that relates to a substantial public health risk. Further, their multidrug resistance added gravity to the antimicrobial resistance (AMR), a globally growing public health threat and this is a critical area of concern especially during the treatment of foodborne gastroenteritis.
Similar content being viewed by others
Introduction
Vibrio parahaemolyticus, a halophile of the genus Vibrio, has emerged globally as one of the leading enteric pathogens associated in food poisoning (toxin-infection), diarrhoea and gastroenteritis resulting from the consumption of raw or under cooked seafood (Nair et al. 2007; Nelapati et al. 2012; Letchumanan et al. 2014). Since early 1996, association of this organism in diarrhoeal cases has witnessed a dramatic increase in the incidence due to emergence and dominance of its unique and highly virulent pandemic O3:K6 serovar (Okuda et al. 1997; Matsumoto et al. 2000; Nair et al. 2007) which is responsible for 50 to 80% of enteric outbreaks in India (Nair et al. 2007; Chowdhury et al. 2013). Still, it is one of the most predominant foodborne pathogens in countries where raw and semi cooked seafood are consumed (Nair et al. 2007). Thermostable direct haemolysin (TDH) and TDH-related haemolysin (TRH) are recognized to be the key virulence determinants of this pathogen and most of the clinical isolates produce either or both of these haemolysins (Tada et al. 1992; Letchumanan et al. 2014). The burgeoning human population invited the growing demand of more food where the aquatic resource including seafood has been considered as a supplementary source of dietary protein. This trend allows the entry of a large number of foodborne pathogens including V. parahaemolyticus in food chain (Austin 2010) resulting the ingestion of contaminated raw or improperly cooked saline water fishes and shellfishes (Nelapati et al. 2012).
In Indian context, most of the studies on this pathogen are mainly concerned with cases of clinical diarrhoea (Chowdhury et al. 2013; Pazhani et al. 2014) and very limited studies have been documented on occurrence of this pathogen in saline water fishes and shellfishes (Deepanjali et al. 2005; Anjay et al. 2014; Guin et al. 2019; Narayana et al. 2020). In our past studies, the occurrence of pathogenic V. parahaemolyticus in crab and shrimp from coastal Odisha and West Bengal (Parthasarathy et al. 2016) and from fish and aquatic environments (Guin et al. 2019) were explored. However, the magnitude of occurrence of pathogenic V. parahaemolyticus in oyster in West Bengal, India is still remained unexplored. It is a growing issue in respect of increasing incidence of V. parahaemolyticus in clinical diarrhoea and gastroenteritis in global context particularly in developing countries including India where marine food source is adopted considerably as a staple alternate dietary protein source (Nair et al. 2007). Further, the recent appearance and dissemination of multiple antibiotic resistance documented among V. parahaemolyticus (Li et al. 2014; Letchumanan et al. 2015; Elmahdi et al. 2016; Ahmed et al. 2018; Narayana et al. 2020) is surely the most remarkable evolution and increasing concern. This draws attention to design the present study to understand the incidence of pathogenic and pandemic V. parahaemolyticus in oysters in coastal part of West Bengal, India as well as to know their potential for transmission to human via food chain by identifying their genetic relatedness with human isolates by PFGE. Also attempted for their antibiotic resistance status in respect to the antimicrobial resistance (AMR), the globally growing issue. Primarily, the generated data will be supportive to highlight the presence of this pathogen in oyster and to generate awareness to the consumers in this region and finally help the seafood industry to strive better by adopting the microbiological food safety measures as well as to facilitate to earn foreign exchange.
Materials and methods
Sample collection and transportation
A total of ninety (90) saline water oysters from Diamond Harbour (52 Km from Kolkata), fishing port of coastal West Bengal, India was collected aseptically in sterile sample container with proper identification label during July 2014 to March 2015 and July 2017 to May 2018 and were transported to the laboratory in the ice container at the earliest and processed immediately.
Enrichment of samples
Isolation of V. parahaemolyticus was performed using the methodology of Kaper et al. (1980). Briefly, with the help of a pair of sterile forceps and scissors, about 10–15 g of gastro-intestinal mass of oyster was properly grinded in sterile pestle and mortar in the vertical laminar flow and transferred the grinded mass in 50 ml capacity test tube containing 30 ml alkaline peptone water (APW) (pre-enrichment broth) and was incubated aerobically for 18–24 h at 37 °C. Sterile forceps and disposable hand gloves were used in handling the collected samples and after processing the forceps and sample containers were properly autoclaved before disposing off.
Isolation of V. parahaemolyticus
Thiosulfate Citrate Bile Salts Sucrose (TCBS) Agar was used as selective media for isolation of V. parahaemolyticus from the collected oyster samples. Briefly, a loop full of overnight grown APW culture was streaked onto the TCBS agar and plates were incubated at 37 °C for 24 h. Five (5) characteristic colonies (typical, round, 2–3 mm in diameter, with green centre) of V. parahaemolyticus on TCBS agar were randomly selected from each plate (represents each sample) for biochemical characterization with Kaper’s multi test medium (Kaper et al. 1980) where the V. parahaemolyticus produce alkali in slant (purple) and acidic (yellow) reaction in butt (K/A). Such positive colonies were considered as presumptive V. parahaemolyticus. Each positive colony was picked up separately with the help of sterile toothpick and was distinctly inoculated on to the nutrient agar plate supplemented with 3.0% NaCl (w/v) as a representative of picked-up colony from TCBS Agar and incubated for 24 h at 37 °C to have a master copy of original culture colony. These presumptive cultures were subjected to species specific Vp-toxR PCR assay for confirmation of species. Disposable hand gloves were used in all the steps and the used gloves & cultured plates were discarded by autoclaving.
Reference strains
Laboratory reference strains Vp-Kx-V138 (Vp-toxR+, tdh+ & GS-PCR+) and VP230 (trh+) were used as positive control in the PCR assays and Escherichia coli K12 strain was used for negative control.
Molecular characterization
Preparation of bacterial lysate for polymerase chain reaction (PCR) assay
A loopful lawn culture of bacterial isolate from LB agar was mixed with 200 μl of sterile water in microcentrifuge tube. This was vortexed for complete mixing and centrifuged at 10,000 rpm for 5 min and boiled for 10 min followed by immediate cooling at 4 °C for 5 min in ice (Kim et al. 1999). The tube was centrifuged at 8000 rpm for 6 min and supernatant was taken as genomic DNA template (bacterial lysate), that was diluted with sterile nuclease free water as required for different PCR method using the set of primer sequence (Table 1).
PCR assay for species confirmation and virulence determination
The toxR (368 bp) is the species‐specific determinative gene sequence for V. parahaemolyticus and all the presumptive isolates were confirmed by identifying this gene sequence by Vp-toxR PCR assay using the methodology of Kim et al. (1999). The Vp-toxR+ isolates were further examined for the presence of virulence gene (tdh) (199 bp), encoded for production of β-haemolysin i.e., TDH, by tdh-PCR assay and trh (250 bp), encoded for TDH related haemolysin, by trh-PCR assay using the methodology of Tada et al. (1992). The tdh+ isolates were subjected to GS-PCR and PGS-PCR assay to determine the presence of pandemic gene sequences by employing the methodology of Matsumoto et al. (2000) and Okura et al. (2004). Bacterial DNA amplification in the aforesaid PCR assays was performed in 25 µl of PCR reaction mixture containing 2.5 µl of 10X PCR amplification buffer, 0.5 µl of dNTP mix (containing 10 mM each, 2 µl (10 pmol/µl) each of forward and reverse primers, 0.2 µl (3 unit/µl) of Taq DNA polymerase, 5.3 µl of bacterial lysate and 14.5 µl of nuclease free water. The amplified DNA fragments were resolved in agarose gel electrophoresis stained with ethidium bromide (0.5 μg/ml) and visualized under UV transilluminator.
Serotyping
Vibrio parahaemolyticus isolates were serotyped using somatic (O) and capsular (K) antisera (Denka Seiken, Co. Ltd., Tokyo) using the methodology of Pazhani et al. (2014).
Pulse field-gel electrophoresis (PFGE)
Pulsed-field gel electrophoresis (PFGE) PFGE was performed according to the PulseNet protocol (Kam et al. 2008). DNA digestion with restriction endonuclease NotI (60 unit/plug; Fermentas, Germany) was performed at 37 °C overnight. Salmonella Braenderup strain H9812 with XbaI digested was used as a molecular weight marker. The restriction fragments were resolved on a CHEF Mapper system (BioRad) in 0.5X Tris–borate-EDTA buffer (TBE) with 300 lM thiourea and the following running conditions: 6 V/cm for 18 h at 14C, with 2–40 s switch time and pump speed of 0.7 L/min. Gels were stained with ethidium bromide (25 min), destained in distilled water for 45 min and photographed under ultraviolet light. The PFGE patterns were analyzed using the BioNumerics version 5.0 software (Applied Maths, Belgium) after normalization of the TIFF images with Salmonella enterica serotype Braenderup size standard. Clustering was performed using the unweighted pair group method (UPGMA) and the Dice correlation coefficient with a position tolerance of 1.5%.
Antibiotic sensitivity test
Antimicrobial susceptibility testing of the pathogenic (tdh+) V. parahaemolyticus isolates were performed using the disk diffusion method with commercially available antibiotic disks such as ampicillin (10 µg), cefotaxime (30 µg), cefpodoxime (10 µg), ceftazidime (30 µg), ceftizoxime (30 µg), ceftriaxone (30 µg), chloramphenicol (30 µg), ciprofloxacin (5 µg), gentamicin (10 µg), levofloxacin (5 µg), nalidixic acid (30 µg), norfloxacin (10 µg), tetracycline (30 µg) and trimethoprim (5 µg) (HiMedia, India) in accordance with the criteria recommended by Clinical and Laboratory Standards Institute (CLSI 2010). Escherichia coli ATCC 25,922 was used as a control strain.
Results
During culture-based isolation using TCBS agar, V. parahaemolyticus colonies were recorded in 88.9% (80/90) of the samples (n = 90). However, on biochemical characterization with Kaper’s multi test medium, 71 of such 80 samples yielded the K/A positive colonies and considered as presumptive V. parahaemolyticus isolates (Table 2). All the presumptive isolates were subjected to Vp-toxR PCR assay for the species confirmation and all were found positive to generate the identifying target 368 bp amplicon; thereby, confirmed as V. parahaemolyticus (Fig. 1A). The confirmed isolates (n = 71) were further screened by tdh-PCR assay using tdh specific primers for the presence of cardinal virulence gene i.e., tdh and 14.08% (10/71) were identified to be virulent by producing specific amplicon (199 bp) (Fig. 1B). However, none of the isolates produced the defined amplicon in trh-PCR assay. Overall, tdh+ V. parahaemolyticus were present in 11.1% (10/90) of the oyster samples. The tdh+ positive isolates (n = 10) were further screened for presence of pandemic genes by group specific PCR (GS-PCR) and pandemic group specific PCR (PGS-PCR) assay using the specific primers described in Table 1. Positive amplicon (235 bp) was detected in 5 isolates with PGS-PCR; however, none in GS-PCR assay (Fig. 1C). Grossly, 5.6% (5/90) of the collected oyster samples were found to harbour V. parahaemolyticus having the pandemic potential.
Amplification of different genes of representative V. parahaemolyticus isolates; A Vp-toxR gene (368 bp), Lane 1 to 5 sample isolates with positive amplicon, M: 100 bp marker, Lane 6: positive control (Vp-Kx-V138), Lane 7: negative control (E. coli K12); B tdh gene (199 bp), Lane 1 to 4 sample isolates with positive amplicon, M: 100 bp marker, Lane 5: positive control (Vp-Kx-V138), Lane 6: negative control (E. coli K12); C PGS gene (235 bp). Lane 1 to 5 sample isolates with positive amplicon, M: 100 bp marker, Lane 6: negative control (E. coli K12), Lane 7: positive control (Vp-Kx-V138)
In the PFGE, two (2) clinical isolates (pandemic serovar) [IDH2916 (O1:K25); IDH4327 (O3:K6)] were grouped in the same clad with 85% similarity in cluster ‘D’ and they were showing ~ 70% relatedness to the oyster isolate (non-pandemic serovar) [(OYS 69 (O9:K17)] belonged to this cluster (Fig. 2). Another human isolate (pandemic serovar) [J23382 (O1: KUT)] showed ~ 62% similarity to the oyster isolate (pandemic serovar) [OYS61 (OUT: K24)] in the cluster ‘A’. The other human isolate [IDH3062 (O4:K8)] alone belonged to cluster ‘E’ and showed the relatedness (~ 53%) to the human and oyster isolates (serovar: pandemic-6 and non-pandemic-1) belonged to the clusters ‘A’, ‘B’, ‘C’ and ‘D’. Further, two oyster isolates [OYS 10 (O10: KUT); OYS 35 (OUT: KUT)] showed > 70% similarity to each other and they revealed > 60% similarity to the other oyster isolate [OYS 79 (O1: KUT)] grouped in the cluster ‘C’. These three oyster isolates (OYS 10; OYS 35 and OYS 79) showed > 55% relatedness to the two human isolates (IDH2916; IDH4327) as well as to one oyster non-pandemic isolate [OYS 69 (O9: K17)] grouped in cluster ‘D’. Two oyster isolates (pandemic serovar) [OYS 43 (O1:K25); OYS 60 (OUT: KUT)] showed ~ 65% relatedness and grouped in cluster ‘B’. Further, these two isolates (OYS 43; OYS 60) exhibited ~ 55% relatedness to another oyster (non-pandemic serovar [OYS 61 (OUT: K24)] and human isolate (pandemic serovar [J23382 (O1: KUT)] belonged to the cluster ‘A’. Besides, these two isolates (OYS 43; OYS 60) found to be related (> 53%) to the human (pandemic serovar) [IDH3062 (O4:K8)] and other oyster isolates of pandemic serovar: [OYS 10 (O10:KUT); OYS 35 (OUT:KUT); OYS 79 (O1:KUT)]; and non-pandemic serovar: [OYS 69 (O9:K17)] belonged to the cluster ‘C’, ‘D’ and ‘E’, respectively (Fig. 2).
Dendrogram of NotI-digested pulsed-field gel electrophoresis (PFGE) profiles with representative of Vibrio parahaemolyticus strains isolated from Oyster in Coastal Bengal, India. Strain J23382, IDH 2916, IDH 3062 and IDH 4327 were the clinical V. parahaemolyticus strains isolated in diarrhoea stools from Infectious Diseases and Beliaghata General Hospital (ID & BG), Kolkata, India
Antibiotic susceptibility analysis showed that all the tdh+ isolates were resistant to cefpodoxime (100%). Ninety percent (90%) of the isolates were resistant to ampicillin and cefotaxime followed by ceftizoxime (60%), tetracycline (50%) and ceftriaxone (40%). On the other hand, all the strains were susceptible to norfloxacin (100%) followed by chloramphenicol and trimethoprim (90% each), ceftazidime and gentamicin (80% each); levofloxacin and nalidixic acid (60% each); ceftriaxone (30%) and tetracycline (10%) (Table 3). Further, 90% of the isolates were found to be intermediately resistant to ciprofloxacin followed by ceftizoxime, levofloxacin & tetracycline (40% each); ceftriaxone & nalidixic acid (30% each); ceftazidime & gentamicin (20% each) and ampicillin, cefotaxime and trimethoprim (10% each).
Discussion
Vibrio parahaemolyticus is a significant foodborne pathogen and causes gastroenteritis through ingestion of contaminated raw or improperly cooked seafoods (Nair et al. 2007). West Bengal, the eastern coastal state of India, is an important center to harvest marine fish and shellfishes. The present study findings revealed that a considerable population (11.1%; 10/90) of oyster in this area were harbouring major virulence gene (tdh+) and have been posing a potential source of pathogenic V. parahaemolyticus. The present data was little higher than the earlier published works in Indian settings where tdh+ was noted in 6.1% and 2.35% isolates in South West coastal region of India (Deepanjali et al. 2005; Narayanan et al. 2020). This variation in frequency may be explained to differences in factors for geographical zones. Moreover, smaller sample size in this study may project for comparatively higher incidence. The observed data were also in support with the findings of previous studies (Hara-Kudo et al. 2003) where tdh+ V. parahaemolyticus was recovered in average of 11% of oyster in Japan. The finding of trh, suggesting the rare possession of trh in population of V. parahaemolyticus from oyster in this area. Further, majority (80%) of the pathogenic (tdh+) isolates belonged to the recognized pandemic serovars (OUT:KUT; OUT:K24; O1:KUT; O1:K25; O10:KUT) that are frequently reported in food borne outbreaks and gastroenteritis in different countries and only two isolates (20%) were of non-pandemic serovars (OUT:K15; O9:K17). Occurrence of pandemic serovars in oyster samples support the trend of prevalence of pandemic serovars in fish and aquatic environments in this area (Han et al. 2017; Guin et al. 2019) and placed the oysters as a potential source of this pathogen for causing food borne problems in the event of taking improperly cooked oysters and its products.
Screening of the pathogenic (tdh+) isolates (n = 10) for their pandemic potential revealed that in PGS-PCR assay, 50% (5/10) isolates were found to harbor the gene of pandemic potential; however, none in GS-PCR. On total sample basis, such pandemic isolates were distributed in 5.5% (10/90) of the oyster samples. The findings reflect the potential of oyster in this area for dissemination of pandemic strains of V. parahaemolyticus and the suitability of PGS-PCR for detecting pandemic strains in samples of saline water origin. The observed results were in concurrence to the findings of previous work by Okura et al. 2004 who concluded that the PGS-PCR assay may be used as a valuable molecular tool not only for detection of pandemic V. parahaemolyticus strains but also for the direct identification of this pathogen polluting the food and environmental samples. The present study findings also supported the observation of Narayanan et al. 2020 who recorded that 14 of 29 (tdh)+ strains were positive in PGS-PCR whereas none in GS-PCR assay. The observed findings indicated that the oyster in this area as a potential source of pathogenic and pandemic V. parahaemolyticus as well as reflected the grade of public health hazard with consumption of improperly cooked oysters. The study reveals the existence of multidrug resistant V. parahaemolyticus in oysters and it was in accord to the findings of earlier published study conducted in Indian settings (Narayanan et al. 2020). The antibiotic cefpodoxime is found to be totally resistant by the isolates. During last few years, frequent use of this antibiotic in treatment of clinical cases including diarrhoea may be the probable cause to acquire such degree of resistance by this pathogen. It was observed that 90% of isolates were resistant to ampicillin which is widely used in aquaculture production systems and the data was supported the earlier studies of Letchumanan et al. (2015); Elmahdi et al. (2016). The findings anticipate the aberrant use of first-generation antibiotics including ampicillin for such growing resistance. Lower resistance was observed against the antibiotics viz. chloramphenicol, gentamicin, levofloxacin, norfloxacin and trimethoprim which is in accord to the earlier study (Letchumanan et al. 2015). This could be attributed to infrequent use of these antibiotics in clinical cases as well as in aquaculture system. Interestingly, 90% of the isolates were found to be intermediately resistant to ciprofloxacin followed by 20 to 40% to ceftazidime, gentamicin, nalidixic acid, levofloxacin, tetracycline and ceftizoxime. It highlighted the trend of growing resistance of this pathogen against a group of antibiotics commonly used in clinical management in Indian context and the magnitude of public health burden in terms of medical cost and antibiotic resistance hazards in very near future.
The phylogenetic analysis demonstrated that irrespective of their serovar belonging i.e., pandemic and non-pandemic, the different isolates from oysters were genetically related in a range of 53 to 70% to the human isolates. The study findings suggest that considerable population of oyster of this study area is the potential source of pathogenic and pandemic V. parahaemolyticus and there is enough possibility for transmission of such V. parahaemolyticus to human being or vice-versa through food chain. Further studies with wider sampling in this aspect may be more supportive to generalize the findings.
In India, the infrastructure along with the physical facilities in most of the fish markets are not adequate (FAO 2001) and V. parahaemolyticus in shellfishes contaminate marketed freshwater fishes. Subsequently, these contaminated market fish may be the source of contamination of other foods in the kitchen and thought to be the probable source of contact of this pathogen in food chain (Sudha et al. 2014). Therefore, generation of awareness on public health and hygiene among the common people will certainly contribute to a great extent to minimize the health problems caused by this pathogen and to lead a healthy community life.
Data availability
The data are original and the work done in the Veterinary Public Health Laboratory of ICAR-Indian Veterinary Research Institute (IVRI), Eastern Regional Station (ERS), Kolkata and at the Bacteriology Laboratory of ICMR-NICED, Kolkata, India.
References
Ahmed HA, El Bayomi RM, Hussein MA, Khedr MHE, Abo Remela EM, El-Ashram AMM (2018) Molecular characterization, antibiotic resistance pattern and biofilm formation of Vibrio parahaemolyticus and V. cholerae isolated from crustaceans and humans. Int J Food Microbiol 274:31–37. https://doi.org/10.1016/j.ijfoodmicro.2018.03.013
Anjay DSC, Kumar A, Kaushik P, Kurmi B (2014) Occurrence of Vibrio parahaemolyticus in marine fish and shellfish. Ind J Geo Marine Sci 43:887–990
Austin B (2010) Vibrios as causal agents of zoonoses. Vet Microbiol 140:310–317. https://doi.org/10.1016/j.vetmic.2009.03.015
Chowdhury G, Ghosh S, Pazhani GP, Paul BK, Maji D, Mukhopadhyay AK, Ramamurthy T (2013) Isolation and characterization of pandemic and non-pandemic strains of Vibrio parahaemolyticus from an outbreak of diarrhea in North 24 Parganas, West Bengal, India. Foodborne Pathog Dis 10:338–342. https://doi.org/10.1089/fpd.2012.1340
CLSI (2010) Methods for antimicrobial dilution and dusk susceptibility testing of infrequently isolated or fastidious bacteria; approved guideline. In: Clinical and laboratory standards institute (ed) M45–2A, 3rd edn. Wayne, PA
Deepanjali A, Kumar HS, Karunasagar I (2005) Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the Southwest coast of India. Appl Environ Microbiol 71:3575–3580. https://doi.org/10.1128/AEM.71.7.3575-3580.2005
Elmahdi S, DaSilva LV, Parveen S (2016) Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: a review. Food Microbiol 57:128–134. https://doi.org/10.1016/j.fm.2016.02.008
FAO (2001) Production, accessibility, marketing and consumption patterns of freshwater aquaculture products in Asia: a cross-country comparison. FAO Fisher Circ 973:283
Guin S, Saravanan M, Anjay CG, Pazhani GP, Ramamurthy T, Das SC (2019) Pathogenic Vibrio parahaemolyticus in diarrhoeal patients, fish and aquatic environments and their potential for inter-source transmission. Heliyon 5(eo1743):1–6. https://doi.org/10.1016/j.heliyon.2019.e01743
Han D, Yu F, Tang H, Ren C, Wu C, Zhang P, Han C (2017) Spreading of Pandemic Vibrio parahaemolyticus O3:K6 and Its Serovariants: a re-analysis of Strains Isolated from Multiple Studies. Front Cell Infect Microbiol 7:188. https://doi.org/10.3389/fcimb.2017.00188
Hara-Kudo Y, Sugiyama K, Nishibuchi M, Chowdhury A, Yatsuyanagi J, Ohtomo Y, Saito A, Nagano H, Nishina T, Nakagawa H, Konuma H, Miyahara M, Kumagai S (2003) Prevalence of thermostable direct haemolysin-producing Vibrio parahaemolyticus O3:K6 in seafood and coastal environment in Japan. Appl Environ Microbiol 69:3883–3891. https://doi.org/10.1128/aem.69.7.3883-3891.2003
Kaper JB, Remmers EF, Colwell RR (1980) A medium for the presumptive identification of Vibrio parahaemolyticus. J Food pro 43:936–938. https://doi.org/10.4315/0362-028X-43.12.936
Kam KM, Luey CK, Parsons MB, Cooper KL, Nair GB, Alam M, Islam MA, Cheung DT, Chu YW, Ramamurthy T, Pazhani GP, Bhattacharya SK, Watanabe H, Terajima J, Arakawa E, Ratchtrachenchai OA, Huttayananont S, Ribot EM, GernerSmidt P, Swaminathan B (2008) Vibrio parahaemolyticus PulseNet PFGE Protocol Working Group. Evaluation and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping Vibrio parahaemolyticus: An international multicenter collaborative study. J Clin Microbiol 46:2766–2773. https://doi.org/10.1128/JCM.00424-08
Kim YB, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M (1999) Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the tox-R gene. J Clin Microbiol 37:1173–1177
Letchumanan V, Chan K, Lee L (2014) Vibrio parahaemolyticus: a review on the pathogenesis, prevalence and advance molecular identification techniques. Front Microbiol 5:705. https://doi.org/10.3389/fmicb.2014.00705
Letchumanan V, Yin WF, Lee LH, Chan KG (2015) Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front Microbiol 6:33. https://doi.org/10.3389/fmicb.2015.00033
Li J, Xue F, Yang Z, Zhang X, Zeng D, Chao G, Jiang Y, Li B (2014) Vibrio parahaemolyticus, Southern Coastal Region of China, 2007–2012. Emerg Infect Dis 20:685–688. https://doi.org/10.3201/eid2004.130744
Matsumoto C, Okuda J, Ishibashi M, Iwanaga M, Garg P, Rammamurthy T, Wong HC, Depaola A, Kim YB, Albert MJ, Nishibuchi M (2000) Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J Clin Microbiol 38:578–585. https://doi.org/10.1128/JCM.38.2.578-585.2000
Nair GB, Ramamurthy T, Bhattacharya SK, Dutta B, Takeda Y, Sack DA (2007) Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin Microbiol Rev 20:39–48. https://doi.org/10.1128/CMR.00025-06
Narayanan SV, Joseph TC, Peeralil S, Mothadaka MP, Lalitha KV (2020) Prevalence, virulence characterization, AMR pattern and genetic relatedness of Vibrio parahaemolyticus isolates from retail seafood of Kerala. India Front Microbiol 11:592. https://doi.org/10.3389/fmicb.2020.00592
Nelapati S, Nelapati K, Chinnam BK (2012) Vibrio parahaemolyticus: An emerging foodborne pathogen—A review. Vet World 5:48–62. https://doi.org/10.5455/vetworld.2012.48-63
Okuda J, Ishibashi M, Hayakawa E, Nishino T, Takeda Y, Mukhopadhyay AK, Garg S, Bhattacharya SK, Nair GB, Nishibuchi M (1997) Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India and isolation of strains from the same clonal group from Southeast Asian travellers arriving in Japan. J Clin Microbiol 35:3150–3155. https://doi.org/10.1128/JCM.35.12.3150-3155.1997
Okura M, Osawa R, Iguchi A, Takagi M, Arakawa E, Terajima J, Watanabe H (2004) PCR-based identification of pandemic group Vibrio parahaemolyticus with a novel group-specific primer pair. Microbiol Immunol 48:787–790. https://doi.org/10.1111/j.1348-0421.2004.tb03596.x
Parthasarathy S, Das SC, Kumar A (2016) Occurrence of pathogenic Vibrio parahaemolyticus in crustacean shellfishes in coastal parts of Eastern India. Vet World 9:330–336
Pazhani GP, Bhowmik SK, Ghosh S, Guin S, Dutta S, Rajendran K, Saha DR, Nandi RK, Mukhopadhyay BMK, Ramamurthy T (2014) Trends in the epidemiology of pandemic and non-pandemic strains of Vibrio parahaemolyticus isolated from diarrheal patients in Kolkata, India. Plos One Neglect Trop Dis 8:2815. https://doi.org/10.1371/journal.pntd.0002815
Sudha S, Mridula C, Selvester R, Hatha AAM (2014) Prevalence and antibiotic resistance of pathogenic Vibrios in shellfish from Cochin market. Indian J Geo- Marine Sci 43:815–824
Tada J, Ohashi T, Nishimura N, Shirasaki Y, Ozaki H, Fukushima S, Takano J, Nishibuchi M, Takeda Y (1992) Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol Cell Probes 6:477–487. https://doi.org/10.1016/0890-8508(92)90044-x
Acknowledgements
The authors are thankful to the Director, ICAR-Indian Veterinary Research Institute (IVRI), Bareilly, U.P. India for providing the necessary funds and facilities for this study. This study was supported in part by the Indian Council of Medical Research (ICMR), Government of India and the Japan Agency for Medical Research and Development (AMED) [Grant Number JP21wm0125004 (to SM)]; Dr. Goutam Chowdhury acknowledges the “Scientist C” position received from the Okayama University, Okayama, Japan.
Funding
The work is done with the fund available in the institute regular grant.
Author information
Authors and Affiliations
Contributions
Planned, designed and supervised: SCD, AKM, AK, SM, SD. Laboratory works: SP, SCD, GC. Analyzed and interpreted the results and compiled the manuscript: SCD, AKM, GC. Finally, all authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Ethical approval
The oyster samples were collected from the retail market with the consent of the retailers. Four (4) clinical archived V. parahaemolyticus strains included for PFGE analysis were received from ICMR-NICED where the samples were routinely referred by the Govt. Hospital and processed following the codal formalities.
Consent for publication
Yes.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Parthasarathy, S., Das, S.C., Kumar, A. et al. Molecular characterization and antibiotic resistance of Vibrio parahaemolyticus from Indian oyster and their probable implication in food chain. World J Microbiol Biotechnol 37, 145 (2021). https://doi.org/10.1007/s11274-021-03113-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-021-03113-3