Abstract
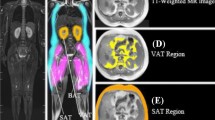
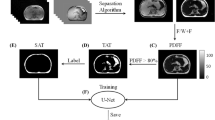
Epicardial adipose tissue is a visceral fat that has remained an entity of concern for decades owing to its high correlation with coronary heart disease. It continues to stump medical practitioners on the pretext of its relevance with pericardial fat and its dependence on a numerous other parameters including ethnicity and physique of an individual. This calls for a fool-proof algorithm that promises accurate classification and segmentation, hence an immaculate prediction. CT is immensely popular and widely preferred for diagnosis. Implementation of an improvised algorithm in CT would be a natural necessity. This research work proposes a Fruitfly Algorithm based Modified region growing algorithm is applied to the acquired CT images to segment fat accurately. The proposed methodology promises image registration and classification in order to segment two cardiac fats namely epicardial, pericardial and mediastinal. The main contributions are (1) Fat feature extraction: Construction of GLCM features CT image (2) Development of GWO based optimal neural network for classification; (3) Modeling the fat segmentation using modified region growing algorithm with Fruitfly optimization. The entire experimentation has been implemented in MATLAB simulation environment and final result is expected to flaunt a definite distinction between cardiac mediastinal and epicardial fats. Parallely, the accuracy, sensitivity, specificity, FPR and FNR have been stated and contrasted methodically with the existing methodology. This venture aims at spurring the healthcare industry towards smarter computational techniques that multiplies efficiency manifold.












Similar content being viewed by others
References
Sicari, R., Sironi, A. M., Petz, R., Frassi, F., Chubuchny, V., De Marchi, D., Positano, V., Lombardi, M., Picano, E., and Gastaldelli, A., Pericardial rather than epicardial fat is a cardiometabolic risk marker: An MRI vs echo study. J. Am. Soc. Echocardiogr. 24(10):1156–1162, 2011.
Sacks, H. S., and Fain, J. N., Human epicardial adipose tissue: A review. Am. Heart J. 153(6):907–917, 2007.
Ding, X., Terzopoulos, D., Diaz-Zamudio, M., Berman, D.S., Slomka P.J., and Dey, D., Automated epicardial fat volume quantification from non-contrast CT. J. Med. Imag., 90340I-9034, 2014.
Pontone, G., Andreini, D., Bertella, E., Petullà, M., Russo, E., Innocenti, E., and Mushtaq, S., Comparison of cardiac computed tomography versus cardiac magnetic resonance for characterization of left atrium anatomy before radiofrequency catheter ablation of atrial fibrillation. Int. J. Cardiol. 179:114–112, 2015.
Oda, S., Utsunomiya, D., Funama, Y., Yuki, H., Kidoh, M., Nakaura, T., and Takaoka, H., Effect of iterative reconstruction on variability and reproducibility of epicardial fat volume quantification by cardiac CT. J. Cardiovasc. Comput. Tomograp. 10(2):150–155, 2016.
Marwan, M., and Achenbach, S., Quantification of epicardial fat by computed tomography: Why, when and how? J. Cardiovasc. Comput. Tomograp. 7(1):3–10, 2013.
Apfaltrer, P., Schindler, A., Schoepf, U. J., Nance, J. W., Tricarico, F., Ebersberger, U., and McQuiston, A. D., Comparison of epicardial fat volume by computed tomography in black versus white patients with acute chest pain. Am. J. Cardiol. 113(3):422–428, 2015.
Aslanabadi, N., Salehi, R., Javadrashid, A., Tarzamni, M., Khodadad, B., Enamzadeh, E., and Montazerghaem, H., Epicardial and pericardial fat volume correlate with the severity of coronary artery stenosis. J. Cardiovasc. Thorac. Res. 6(4):235–239, 2014.
Yalamanchilia, R., Dey, D., Kurkure, U., Nakazato, R., Berman, D. S., and Kakadiaris, I. A., Knowledge-based quantification of pericardial fat in non-contrast CT data. Proc. SPIE 7623:76231X–762317X.
Rodrigues, É. O., Conci, A., Morais, F. F. C., and Pérez, M. G., Towards the automated segmentation of epicardial and mediastinal fats. 2015 Elsevier Ireland Ltd. All rights reserved.
Lin, D.-T., Yan, C.-R., and Chen, W.-T., Autonomous detection of pulmonary nodules on CT images with a neural network-based fuzzy system. J. Comput. Med. Imag. Graph. 29(6):447–458, 2005.
Ling, Z., McManigle, J., Zipunnikov, V., Pashakhanloo, F., Khurram, I. M., Zimmerman, S. L., and Philips, B., The association of left atrial low-voltage regions on electro anatomic mapping with low attenuation regions on cardiac computed tomography perfusion imaging in patients with atrial fibrillation. J. Heart Rhythm 12(2):857–864, 2015.
Isgum, I., Staring, M., Rutten, A., Prokop, M., Viergever, M. A., and Van Ginneken, B., Multi-atlas-based segmentation with local decision fusion—Application to cardiac and aortic segmentation in CT scans. IEEE Trans. Med. Imaging 28(7):1000–1010, 2009.
Rodrigues, É. O., Morais, F. F. C., Morais, N. A. O. S., Conci, L. S., Neto, L. V., and Conci, A., A novel approach for the automated segmentation and volume quantification of cardiac fats on computed tomography. Comput. Methods Prog. Biomed. 123:109–128, 2016.
Champier, J., Cinotti, L., Bordet, J.-C., Lavenne, F., and Mallet, J.-J., Delineation and quantization of brain lesions by fuzzy clustering in positron emission tomography. J. Comput. Med. Imaging Graph. 20(1):31–41, 2006.
Bandekar, A. N., Naghavi, M. and Kakadiaris, I. A., Automated pericardial fat quantification in CT data. Proceedings of the 28th IEEE EMBS annual international conference new York City, USA, 2006.
Guo, Z., Xin, Y., Liu, S., Lv, X. and Li, S., Comparisons of fat quantification methods based on MRI segmentation. IEEE, 978–1–4799-3979-4/14/$31.00, 2014.
Zlokolica, V., Velicki, L., Janev, M., Mitrinovic, D., and Babin, D., Epicardial fat registration by local adaptive morphology-thresholding based 2D segmentation. IEEE, 2014.
Sumitpaibul, P., Damrongphithakkul, A., and Watchareeruetai, U., Fat detection algorithm for liver biopsy images. IEEE, 978–1–4799-3174-3/14/$31.00, 2014.
Antony, J., McGuinness, K., Welch, N., Coyle, J., and Franklyn-Miller, A., Fat quantification in MRI-defined lumbar muscles. IEEE, 978–1–4799-6463-5/14/$31.00, 2014.
Yan, Z., Tan, C., Zhang, S., Zhou, Y., and Belaroussi, B., Automatic liver segmentation and hepatic fat fraction assessment in MRI. IEEE, 1051–4651/14 $31.00, 2014.
Ahmed, S., Gilles, B., Puech, W., Hassouni, M. E and Rziza, M., Hierarchical MRI segmentation of the musculoskeletal system using texture analysis and topological constraints. 978–1–4799-4572-6/14/$31.00, 2014.
Song, H., Zhang, Q., Sun, F., Wang, J., and Wang, Q., Breast tissue segmentation on MR images using KFCM with spatial constraints. IEEE, 978–1–4799-5464-3/14/$31.00, 2014.
Ahmad, E., Yap, M. H., Degens, H. and McPhee, J., Enhancement of MRI human thigh muscle segmentation by template-based framework. IEEE, 978–1–4799-5686-9/14/$31.00, 2014.
Spasojeviü, A., Stojanov, O., Turukalo, T. L., and Šveljo, O., Estimation of subcutaneous and visceral fat tissue volume on abdominal MR images. IEEE, 978–1–4799-5888-7/14/$31.00, 2014.
Ayerdi, B., Echaniz, O., Savio, A., and Graña, M., Automated segmentation of subcutaneous and visceral adipose tissues from MRI. Springer. 427–433, 2016.
Brinkley, T. E., Tina, E., Hsu, F.-C., Jeffrey Carr, J., Gregory Hundley, W., Bluemke, D. A., Polak, J. F., and Ding, J., Pericardial fat is associated with carotid stiffness in the multi-ethnic study of atherosclerosis. J. Nutr. Metab. Cardiovasc. Dis. 21(5):332–338, 2011.
Bandekar, A. N., Naghavi, M., and Kakadiaris, I. A., Automated pericardial fat quantification in CT data. Eng. Med. Biol. Soc. Int. Conf. IEEE. 932–935, IEEE, 2006.
Kaus, M. R., von Berg, J., Weese, J., Niessen, W., and Pekar, V., Automated segmentation of the left ventricle in cardiac MRI. J. Med. Image Anal. 8(3):245–254, 2004.
Premkumar, R., and Anand, S., Secured and compound 3-D chaos image encryption using hybrid mutation and crossover operator. Multimed. Tools Applic., 2018. https://doi.org/10.1007/s11042-018-6534-z.
Mangili, L. C., Mangili, O. C., Bittencourt, M. S., Miname, M. H., Harada, P. H., Lima, L. M., Rochitte, C. E., and Santos, R. D., Epicardial fat is associated with severity of subclinical coronary atherosclerosis in familial hypercholesterolemia. J. Atherosclero. 254:73–77, 2016.
Meenakshi, K., Rajendran, M., Srikumar, S., Chidambaram, S., Epicardial fat thickness: A surrogate marker of coronary artery disease – Assessment by echocardiography. 2015 cardio logical Society of India, published by Elsevier B. V, All rights reserved.
Sicari, R., Sironi, A. M., Petz, R., Frassi, F., Chubuchny, V., Marchi, D.D., Positano, V., Lombardi, M., Picano, E., and Gastaldelli, A., Pericardial rather than Epicardial fat is a Cardiometabolic risk marker: An MRI vs Echo study. J. Am. Soc. Echocardiogr., 24(10).
Yang, F., Ding, M., Zhang, X., Hou, W., and Zhong, C., Non-rigid multi-modal medical image registration by combining L-BFGS-B with cat swarm optimization. J. Inf. Sci. 316:440–456, 2015.
Guo, Z., Xin, Y., Liu, S., Lv, X. and Li, S., Comparisons of fat quantification methods based on MRI segmentation. IEEE, 978–1–4799-3979-4/14/$31.00, 2014.
Bucher, A. M., Joseph Schoepf, U., Krazinski, A. W., Silverman, J., Spearman, J. V., De Cecco, C. N., Meinel, F. G., Vogl, T. J., and Geyer, L. L., Influence of technical parameters on epicardial fat volume quantification at cardiac CT. Eur. J. Radiol. 84(6):1062–1067, 2015.
Premkumar, R., and Anand, S., Secured permutation and substitution based image encryption algorithm for medical security application. J. Med. Imag. Health Inform. 6(8):2012–2018, 2016.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Image & Signal Processing
Rights and permissions
About this article
Cite this article
Priya, C., Sudha, S. Adaptive Fruitfly Based Modified Region Growing Algorithm for Cardiac Fat Segmentation Using Optimal Neural Network. J Med Syst 43, 104 (2019). https://doi.org/10.1007/s10916-019-1227-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-019-1227-3