Abstract
This study investigates the influence of C3 and C4 plants, soil texture and seasonal changes on the structure and assimilation of plant-derived C of soil microbial communities. In 2012 we collected soil samples in the growing and non-growing season from a vegetation change experiment cropping herbaceous C3 and C4 plants for 6 years on two soils differing in their texture. Phospholipid fatty acids and their compound-specific δ13C values were used to determine microbial community biomass and its composition and the assimilation of C from plants to soil microorganisms, respectively. While soil microbial biomass differed mainly between seasons, the microbial community composition was related to soil texture. The proportion of plant-derived C assimilated by soil microorganisms was best explained by soil texture, too. In contrast, differences in photosynthetic pathways of plants had no impact on microbial biomass or on microbial community composition but expectedly on the isotopic composition of the microbial markers. Our results demonstrated that vegetation, differing in C3 plants and C4 plants, has no effect on the soil microbial community and their proportion of assimilated C derived from plants, if plants are similar in their productivity and phenology. Thus, our study verifies that vegetation change experiments are beneficial in exploring the interactions of plant soil and microbes and how environmental properties, such as seasonality or soil type impact this interaction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Understanding the factors that govern soil carbon (C) dynamics, such as the accumulation and decomposition of soil organic matter (SOM), is critical as soils store most of the terrestrial organic C (Lal 2004) and therefore are an important component in the global C cycle. Soil microorganisms play a key role in controlling formation, decomposition and accumulation of SOM (Balser and Firestone 2005; Cotrufo et al. 2015; Gleixner et al. 2002; Lange et al. 2015; Liang et al. 2017). Although the soil microbial activity is influenced by several factors, such as soil temperature, soil humidity or pH (Voroney and Heck 2015), it is largely controlled by plant-derived C resources (Wardle 2002). In turn, microbial activity remobilizes nutrients that enhance plant growth (Porazinska et al. 2003). In addition, the soil microbial community composition is strongly affected by abiotic and biotic factors such as soil texture (de Vries et al. 2012; Johnson et al. 2003; Merckx et al. 1985), pH (Thoms et al. 2010), soil moisture (Berg and Steinberger 2010; Lange et al. 2014), temperature (Medeiros et al. 2006), seasonality (Cao et al. 2011; Habekost et al. 2008), and vegetation type (Berg and Steinberger 2010; Epron et al. 2011; Grayston et al. 2001). However, little is known about how changes in the composition of the microbial community affect ecosystem functions. Therefore, it is important to know the relative importance and possible interactions between environmental factors in shaping the soil microbial community and ultimately the ecosystem functions, in particular those of soil C dynamics.
Microbial communities in soils are commonly studied by analyses of phospholipid fatty acids (PLFA; Frostegard et al. 1991; Frostegård et al. 2011; Zelles 1997, 1999). The PLFA method is particularly advantegeous when working with natural isotopic abundances as it is highly sensitive to the shifts in the isotopic signatures of lipid biomarkers (Pett-Ridge and Firestone 2017). Other techniques may be more appropriate if high isotope label is used, for instance stable isotope probing is increasingly used combining high isotope label with molecular techniques that allow a finer taxonomic resolution of microbial communities than the PLFA technique (Hungate et al. 2015; Mau et al. 2015). The information gained by the PLFA method is comparable to the ecological concept of functional groups (Joergensen and Wichern 2008) and is particularly useful when investigating soil C dynamics. Specific PLFA markers might be assigned to distinct microbial groups in soils: Gram-positive (G+) bacteria, which are commonly considered as decomposers of SOM (Bahn et al. 2013; Mellado-Vázquez et al. 2016); Gram-negative (G−) bacteria are supposed to have high affinity for plant-derived C, such as root exudation (Denef et al. 2009; Mellado-Vázquez et al. 2016); and saprotrophic fungi which are able to decompose root exudates and plant litter (Treonis et al. 2004) as well as SOM (Garcia-Pausas and Paterson 2011; Mellado-Vázquez et al. 2016).
A common method for a better understanding of C flow between plants, microorganisms and SOM are vegetation change experiments (e.g. Balesdent and Balabane 1996; Gleixner 2013), which in combination with PLFA and stable isotope ratios analyses (Evershed et al. 2006; Garcia-Pausas and Paterson 2011; Kindler et al. 2009) have been used to explore interactions among plants, soil and microorganisms (Pett-Ridge and Firestone 2017). In such experiments, the native plant community of either C3 or C4 plants is exchanged by the respective other plant type, while the soil remains the same, and receives the isotopic imprint of the other plant type over time. Differences in the photosynthetic pathways of C3 and C4 plants result in naturally distinct δ13C values of both plant types (C3: approx. − 25‰; C4: approx. − 12‰; Degens 1969; O’Leary 1981). The assimilation of plant-derived carbon during microbial growth and biomass formation, including lipid synthesis, translates the differences in δ13C values between the two plant types into microbial PLFAs. In the course of the lipid synthesis an isotopic fractionation takes place, which reduces the δ13C values of microbial lipids by 6‰ to 8‰ compared to the carbon sources (DeNiro and Epstein 1977). However, there are uncertainties if such a vegetation change from C3 to C4 plants only changes the isotopic signal of the C source or if it also causes changes in the abundance and composition of the soil microbial community. Furthermore the photosynthetic pathway of plants itself has been suspected to impact soil carbon dynamics (Chen et al. 2016). This would indicate that results obtained in such experiments are confounded. Thus, it is important to assess the relative importance of environmental factors, such as soil texture and season compared to the vegetation change affecting the soil microbial community.
In this study we investigated the effect and the relative importance of a C3–C4 vegetation change, soil properties and season on total soil microbial biomass, soil microbial community composition as well as their isotopic C composition in a replicated C3–C4 vegetation change experiment. Specifically we asked if (i) the soil microbial community composition and abundance are affected by photosynthetic pathway, soil properties and seasonal changes, (ii) how these factors influence the isotopic values of microbial community and their proportion of assimilated C from plants, and (iii) if the assimilation of plant-derived C differs among microbial groups. Therefore, C3 and C4 plants, similar in their phenology and productivity, were grown for six years on two soils differing in texture, pH and organic carbon content at the same site with the same climatic conditions. Thus, the influence on the experimental treatment by uncontrolled conditions could be minimized. Finally, we calculated the microbial assimilation of plant-derived C for distinct microbial groups depending on soil properties and season.
Materials and methods
Site description and experimental design
The C3–C4 vegetation change experiment was established at the Max Planck Institute for Biogeochemistry in Jena, Germany in 2002. Four plots of 24 m2 were set up directly next to each other to avoid environmental biases, such as climate. Two different types of homogenized soil were filled into two plots each to a depth of 2 m. Both soils differed in their texture and pH. The first one was originally derived from a forest A-horizon and had a coarse texture (50% sand, 44% silt and 6% clay; pH 6.9) and relatively higher soil organic carbon (SOC) concentration (Table 1) and is henceforth termed as “coarse soil”. The second soil was derived from the B-horizon of a calcareous soil and had finer texture (9% sand, 75% silt and 16% clay; pH 7.8) but lower SOC concentration (Table 1), and is henceforth termed as “fine soil”. Both soils originally had C3 vegetation and until 2006 the entire experiment was continuously cropped with C3 vegetation (Malik et al. 2012; Scheibe et al. 2012). The selection of plant species was based on comparable biomass production and phenology. Scorpion weed (Phacelia tanacetifolia Benth.), sunflower (Helianthus annuus L.) and wheat (Triticum spec. L.) were rotationally grown on all plots as two-species community. In November 2006 on one plot with coarse soil and one plot with fine soil the vegetation change started, while on the other plots with coarse and with fine soil were continuously cropped with C3 plants. The rotationally grown C4 plant species were lovegrass (Eragrostis curvula Wolf), maize (Zea mays L.), amaranth (Amaranthus spec. L.) and sorghum (Sorghum spec. Moench). All plots were equally managed, namely annually in autumn the total shoot biomass was harvested and equal amounts were re-distributed on each plot considering the vegetation type. From October to April, i.e. during the non-growing season, plots were covered with a water permeable sheet that allowed plant litter decomposition but prevented weed germination. In the following spring the plots were sown again with their respective vegetation type.
Soil sampling
Soil samples were collected in 2012 in February (non-growing season) and June (growing season). Using a stainless steel core cutter (inner Ø = 4.8 cm, Eijkelkamp Agrisearch Equipment, Giesbeek, The Netherlands, 0–10 cm deep), three independent samples were collected along the entire length of one side of each plot. Soil samples were sieved (< 2 mm) and cleaned by removing remaining plant material and stones. Clean and sieved soil samples were stored at − 20 °C for less than a week before PLFA extraction, and the extracts were subsequently stored at − 20 °C for less than a month before analysis.
PLFA extraction, analysis and compound specific measurement
A mixture of CH3OH, CHCl3 and 0.05 M K2HPO4 buffer (2:1:0.8 v/v/v) was used to extract the total lipid fraction from 50 g of wet soil (Bligh and Dyer 1959; modified by Kramer and Gleixner 2006). PLFAs were purified from the total lipid extraction by sequentially eluting a silica-filled solid phase extraction column (Bond Elut® SPE, Varian US.) with chloroform, acetone and methanol; the methanol elution, which contained the PLFA fraction, was hydrolyzed and methylated with a 0.2 M KOH methanolic solution. Subsequently, methylated fatty acids (FAMEs) were split up into saturated (SATFA), monounsaturated (MUFA) and polyunsaturated fatty acids (PUFA) in an aminopropyl modified SPE column (Bond Elut® SPE, Düren, Germany) impregnated with AgNO3. After adding the FAME 19:0 (as an internal standard) to all fractions, FAMEs were quantified with a GC-FID system (GC: HP 6890 Series, AED: G 2350 A, Agilent Technologies, United States) using a HP Ultra column (50 m × 0.32 mm internal diameter, 0.52 mm film thickness) and Helium as a carrier gas. Following the initial conditions, 140 °C held for 1 min, the temperature increased at a rate of 2° min−1 until reaching 270 °C (6 min isotherm). Afterwards, the heating rate increased to 30 °C min−1 to reach a final temperature of 320 °C that was held during 3 min. PLFA identification was performed using GC/MS (Thermo Electron, Drereich, Germany) by comparison of PLFA peak retention times and their mass spectra with known standards and mass spectral data from an in house database (Thoms et al. 2010; Thoms and Gleixner 2013).
PLFA nomenclature of individual markers is used as follows: the number of C atoms is followed by a colon and the number of double bonds. The position of the first double bond is denoted by the symbol omega (ω) and the number of C atoms from the aliphatic end of the molecule (e.g.: 16:1ω5), if there are multiple double bonds, the position of the subsequent double bond are indicated after a coma following the first one (e.g.: 18:2ω6,9). Methyl branching at the iso, anteiso and tenth C from the carboxylic end is represented by the prefixes “i” (e.g.: i15:0), “a” (e.g.: a16:0) and “10Me” (e.g.: 10Me16:0), respectively. The prefix “cy” denotes cyclopropane fatty acids (e.g.: cy19:0). Based on their predominant microbial origin, individual PLFA markers were assigned to different microbial groups. PLFA markers assigned for Gram-positive (G+) bacteria were i15:0, a15:0, i16:0, i17:0 and a17:0 (Zelles 1997). The markers 10Me16:0, 10Me19:0 were assigned to actinobacteria (Kroppenstedt 1985), which is a subgroup of the G+ bacteria. PLFA markers assigned for Gram-negative (G−) bacteria were 15:1, 16:1ω7, 16:1ω5, 16:1, 17:1, 18:1ω9, 18:1ω7, cy17:0 and cy19:0 (Zelles 1997). The cyclic markers cy17:0 and cy19:0 are produced under environmental stress conditions by G− bacteria (Kaur et al. 2005). However it has been observed that cyclic PLFA markers were predominately observed in environments that are richer in G+ bacteria than G− bacteria (Mellado-Vázquez et al. 2016; Treonis et al. 2004). Therefore, we consider cyclic markers as a separate marker group (“cy G− bacteria”) from non-cyclic markers assigned to G− bacteria. Finally, only the PLFA marker 18:2ω6,9 was used as proxy for saprotrophic fungi and their biomass (Frostegard et al. 2011). As proxy for the total microbial biomass, the concentrations of all PLFA markers were summed, including the non-specific markers 14:0, 16:0 and 18:0 (Zelles 1997).
Carbon isotope ratios of individual PLFA markers were measured in triplicate in a GC-IRMS system (HP5890 GC, Agilent Technologies, Palo Alto USA; GC combustion III and IRMS: Deltaplus XL, Finnigan MAT, Bremen, Germany), using a HP Ultra column (50 m × 0.32 mm internal diameter, 0.52 mm film thickness) and helium as a carrier gas. The FAME 19:0 was used as internal standard to assess measurement precision (mean reproducibility of 0.23‰, n = 72) and for the offset correction (Werner and Brand 2001). A SATFA-mix, was injected as external standard before each triplicate sample measurement and fatty acid 19:0 was used for drift correction (Werner and Brand 2001). The δ13C values of SATFA were analyzed with split mode (1:10); whilst δ13C values of MUFA and PUFA were measured with splitless mode. The initial oven temperature of 140 °C was held for 1 min, followed by an increase in temperature at a rate of 2 °C min−1 until reaching 252 °C. Followed by a heating rate of 30 °C min−1 until a final temperature of 320 °C that was held during 3 min. The software ISODAT NT 2.0 (SP 2.67, Thermo Fisher, USA) was used for data evaluation. Isotope ratios are expressed as δ13C value in per mil (‰) relative to the international reference standard Vienna-PeeDee Belemnite (V-PDB) (Eq. 1) using NBS 19 (Werner and Brand 2001):
where (13C/12C)sa is the 13C/12C ratio of the sample and (13C/12C)std the 13C/12C ratio of the reference standard V-PDB. δ13C values were also corrected for the methyl carbon added during methylation (Eq. 2; Kramer and Gleixner 2006):
where δ13CPLFA is the isotope ratio of the phospholipid fatty acid, δ13CFAME the isotope ratio of the phospholipid fatty acid methyl ester, δ13CMeOH that of methanol used for derivatization and NPLFA is the number of carbon atoms of the PLFA.
Assessing the C origin in PLFA markers
The net proportion of assimilated C derived from plants to individual PLFAs compared to soil-derived C (FpPLFA; Eq. 3) was calculated according to Kramer and Gleixner (2006):
where δ13CPLFA–C4 and δ13CPLFA–C3 represent the isotopic values of individual PLFA markers collected simultaneously in soils with C4 and C3 vegetation. The isotopic values of both vegetation types analyzed in the year of sampling are represent by δ13CPlant–C4 and δ13CPlant–C3 (Table 1). Bulk samples of all plant communities were dried at 70 °C for 48 h and ground with a ball mill prior to chemical analysis. The carbon isotopic composition of dried samples was analyzed using a DeltaPlus isotope ratio mass spectrometer (Thermo Fisher, Bremen, Germany) coupled via a ConFlowIII open-split to an elemental analyzer (Carlo Erba 1100 CE analyzer; Thermo Fisher Scientific, Rodano, Italy).
Explanatory variables
In addition to the experimental design variables (vegetation type, soil texture and season) ecologically important covariates were assessed. Soil moisture (%) was determined gravimetrically (Black 1965) from 5 g of soil (wet weight) that were collected as subsamples from the soil cores taken for PLFA analysis. Root biomass (g m2) was quantified as the average of roots collected from three squares (0.5 × 0.5 m) in each plot. In each square a soil core was taken (4.8 cm in diameter, 0–10 cm deep) using a split tube sampler to obtain SOC content. After sampling, the soil was dried at 40 °C until constant weight and homogenized by grinding in a ball mill. Concentration of SOC was calculated from the difference between total C and inorganic C. Total C and inorganic C were measured by elemental analysis at 1150 °C (Vario Max; Elementar Analysensysteme GmbH, Hanau, Germany), inorganic C was obtained after burning organic C for 16 h at 450 °C in a muffle furnace (Steinbeiss et al. 2008).
Statistical analysis
Linear mixed-effects models (LMM) were applied to test for effects of the experimental factors (photosynthetic pathways, soil texture and season) on the response variables (microbial biomass, weighted average of δ13C values of PLFA markers and weighted average of the proportion of assimilated C from plants per plot). To account for the repeated measurements “plot” was fitted as random factor. For analyzing the response of the specific soil microbial groups, all respective PLFA markers were considered individually to account for e.g. differences in the synthesis among markers resulting in different δ13C values. Therefore, “plot” and “PLFA marker” were fitted as random factors in these LMM. Starting from a constant null model, the factor “photosynthetic pathways of plants” was fitted first, followed by soil texture, season and the interaction terms among each other. The maximum likelihood method was used and likelihood ratio tests were applied to assess the statistical significance of model improvement (Zuur et al. 2009). The importance of the factors was assessed on the basis of the proportion of the additionally explained variance (R2) in the sequential models comparison, using the “r.squaredGLMM” function in MuMIn package in R. The effect and the importance (R2) of the experimental factors on the microbial community composition based on relative PLFA concentrations were tested by Permutational Multivariate Analyses of Variance (PERMANOVA: “adonis” function in vegan package in R (Oksanen et al. 2011). For PERMANOVAs the “Bray–Curtis” distance measure was applied. In addition, redundancy analyses (RDAs) were carried out to relate the effects of experimental factors on the soil microbial community composition to other environmental parameters by assessing the impact of root biomass, SOC and soil moisture. Permutations for hierarchical design were selected to account for the replicated sampling. RDAs were carried out using CANOCO 5.0 for windows (ter Braak and Šmilauer 2012). General markers were not considered performing PERMANOVAs and RDAs.
Results
Microbial biomass and microbial community composition
Microbial biomass (measured as total PLFA concentration) was significantly impacted by season and soil texture, which explains 61% and 9% respectively of the variations (Fig. 1a, Table 2). More microbial biomass was observed in the growing season and in fine soils. Photosynthetic pathway had no impact on soil microbial biomass (Fig. 1a, Table 2). However, the analyses of individual markers showed that the concentration of PLFA markers differed strongly among the microbial groups (Table S1). Season, but not soil texture, also had a significant effect on the concentrations of individual PLFA markers and differed among microbial groups as shown by the significant interaction term between microbial group and season (Table S1a). This significant interaction term indicates that the biomass of microbial groups differently changes in the course of the year. In particular PLFAs markers assigned to G− bacteria were much higher in concentration in the growing season than in the non-growing season, while the concentration of the other PLFA markers did not change to that extent (Fig. 1b).
Total microbial biomass (a) in coarse and fine soils sown with C3 and C4 plants in different seasons (non-growing and growing), and the microbial biomass of different microbial groups (b) in the non-growing and growing seasons in coarse and fine soils. Error bars represent the standard error of mean (SEM)
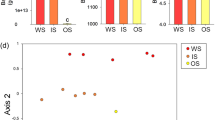
Soil texture explained 43% of total variation of the microbial community composition (Table 2). The photosynthetic pathway and season did not affect the composition of the soil microbial community. The RDA revealed that the significant effect of soil texture was accompanied by differences in SOC content and root biomass (Fig. 2). Due to collinearities among both environmental variables, only the first variable chosen in the forward selection was significant. Together, SOC and root biomass account for 45.1% of the variance in PLFA composition. On plots of our study more root biomass was found in fine soils, while in coarse soils SOC content was higher (Table 1). The RDA further revealed that root biomass and SOC content mainly separates G+ bacteria and cy G− bacteria markers from G− bacteria markers (Fig. 2).
Triplot showing the results of a redundancy analysis (RDA) of the mean relative proportions (mol%) of individual phospholipid fatty acids (PLFAs) in coarse and fine soils sown with C3 and C4 plants. Root biomass (RB), soil organic carbon (SOC) and soil moisture were set as explanatory variables to explain the variance of individual PLFAs. Stars indicate significant effects of explanatory variables (p < 0.05). The microbial groups to which the PLFA markers are assigned are given in square brackets: Gram-positive bacteria [G+], actinobacteria [Actino], cy Gram-negative bacteria [G− cy], Gram-negative bacteria [G −] and saprotrophic fungi [SF]. For simplification reasons, the graph shows average plot values, while the statistics presented in Table 3 are based on all replicates per plot
Isotopic signature of microbial markers and assimilation of plant-derived C
The plant photosynthetic pathway explained the δ13C values of the microbial community (weighted plot average of PLFAs) by 91% (Fig. 3a, Table 2). As expected, all individual PLFAs had higher δ13C values on C4 vegetated plots (av = − 21.6‰ ± 3.2 SD) compared to C3 vegetated plots (− 28.8‰ ± 2.6; individual markers are shown in Table S2). However, the δ13C enrichment on C4 plots differed among microbial groups and season (Fig. 3b, Table S1b). During the growing season, an increase of ~ 2‰ of δ13C values in PLFA markers indicating G+ bacteria, actinobacteria and cy G− bacteria (i15:0, i16:0, i17:0, a17:0, 10Me16:0, 10Me19:0, cy17:0 and cy19:0) was observed compared to the values observed in the non-growing season (Table S2a, b, c). In contrast, not all the δ13C values of PLFA markers indicative for G− bacteria were affected the season; δ13C values of 15:1, 16:1ω5 and 18:1ω9 were higher in the non-growing season (Table S2d), while the other G− bacteria markers (16:1ω7, 16:1, 17:1, 18:1ω7) showed no difference of their δ13C values between seasons.
Isotopic values of the soil microbial community as (a) weighted average per plot in coarse and fine soils sown with C3 and C4 plants in different seasons (non-growing and growing), and of microbial groups (b) on C3 and C4 vegetated plots in coarse and fine soils. Error bars represent the standard error of mean (SEM)
The portion of plant-derived C in the entire soil microbial community (weighted plot average of PLFAs) was not different between soil types (Table 2, Fig. 4a). However, we found specific differences in the C assimilation patterns among microbial groups (Fig. 4b, Table S1c), with highest portions in saprotrophic fungi (73.3% ± 13.0) and G− bacteria (53.8% ± 15.0). Furthermore, the assimilation of plant-derived C among microbial groups was differently affected by season and soil texture (Fig. 4b, Table S1c). The assimilation of plant-derived C in G− bacteria was higher in fine soils (58.0% ± 19.5) than in coarse soils (49.6% ± 8.3). In contrast, plant-derived C was on average higher in the coarse soil than in fine soil in PLFAs assigned for G+ bacteria (coarse soil: 51.3% ± 2.9, fine soil: 40.8% ± 9.1), actinobacteria (45.5% ± 6.3, 36.4% ± 18.4) as well as in cy G− bacteria markers (49.5% ± 11.0, 41.4% ± 7.3). Saprotrophic fungi hold more plant-derived C in the non-growing season (83.1% ± 9.6) than in the growing season (63.4% ± 6.7) as well as G− bacteria (60.3% ± 11.8, 47.4% ± 15.8). In contrast, plant-derived C was similar between seasons in PLFAs assigned for G+ bacteria (growing season: 47.5% ± 10.5, non-growing season: 44.7% ± 6.6), actinobacteria (38.1% ± 7.1, 43.9% ± 18.9, %) and cy G− bacteria markers (46.8% ± 7.0, 44.2% ± 12.7, %, Fig. 4b).
Discussion
We found no differences in the microbial biomass and in the microbial community composition between different photosynthetic pathways. In our experiment, the C3 and C4 plant communities were similar in biomass production and phenology, and thus the photosynthetic pathway alone has no or at most a minor impact on soil microbial communities and their assimilation of plant-derived carbon. This finding suggests that impact of different photosynthetic pathways on the microbial community (e.g. Chen et al. 2016), might be triggered by confounding effects of other environmental parameters.
We found a general increase in the total microbial biomass in the growing season, which is most probably caused by higher temperatures and increased availability of C resources during vegetation periods, e.g. root litter and exudates. In contrast, seasonal changes did not have an impact on the composition of the different bacterial groups. This indicates that the soil bacterial groups responded similarly to environmental changes accompanied with the seasonal variation. The similar response might be caused by an adaptation of the soil microorganisms during the 6 years of cultivation prior to sampling. Furthermore, the total soil microbial biomass depended on soil type and was significantly higher in soils of fine grain size. In addition the abundance of bacterial groups was differently affected by soil types. PLFA markers assigned to G− bacteria were more abundant in fine soils. In our experiment more root biomass was found in fine soils, which form a finer texture and might therefore sustain a better root net resulting in more root biomass than in coarse soils (Merckx et al. 1985). Thus, the higher abundance of G− bacteria in the soil of fine soil might be mediated by the higher root biomass. In contrast, higher proportional abundances of G+ bacteria, actinobacteria and cy G− bacteria were mainly found in the coarse soil and may be related to the higher SOC content in the coarse soil of our experiment. These microbial groups are suggested to be better adapted than other microbial groups to degrade SOM (Fierer et al. 2003) and to live on these spatially dispersed C sources (Don et al. 2013). However, the parameters of the selected soil types are inter-correlated in this study and therefore the points discussed above are suggested as possible mechanisms underlying the observed effects of the two soil types on microbial biomass and microbial composition.
The portion of plant-derived C contained in the soil microbial community was not different between soil types (Table 2, Fig. 4a). However, the specific differences in the C assimilation patterns among microbial groups in different soil type (Fig. 4b, Table S1), point to an impact of environmental factors on C utilization strategies among microbial groups.
The plant-derived C assimilation by bacterial groups (G+ bacteria, actinobacteria, cy G− bacteria, G− bacteria) was similar among seasons. In contrast, saprotrophic fungi took up more plant derived C in the non-growing season than in the growing season. In the non-growing season, when the most important plant C resource was derived from decaying plant biomass, the amount of plant C incorporated into saprotrophic fungi was significantly higher compared to the other microbial groups. This is likely the result of a relative advantage for the fungi in degrading litter (Klamer and Hedlund 2004; Singh et al. 2006).
As expected, the δ13C values of the PLFAs were primarily driven by the photosynthetic pathway of plants, namely higher δ13C values in C4 growing plots than in C3 growing plots reflected the naturally higher δ13C values of C4 plants. However, the δ13C values of some bacterial PLFA markers differed unexpectedly between the non-growing and the growing seasons. PLFA markers, assigned to soil associated bacteria (G+ bacteria: i15:0, i16:0, i17:0, a17:0; actinobacteria: 10Me16:0, 10Me19:0; cy G− bacteria: cy17:0, cy19:0; Mellado-Vázquez et al. 2016) were enriched by 2‰ in the δ13C values in the growing season compared to the non-growing season. This enrichment might reflect an increased microbial respiration and higher C turnover with increasing soil temperature (Creamer et al. 2015), but also they maybe had access to more enriched plant-derived C sources. Contrarily, some G− bacterial PLFA markers (15:1, 16:1ω5 and 18:1ω9), showed a higher enrichment in their δ13C values as well as higher portions of plant-derived C in the non-growing season than in the growing season. This finding was surprising as G− bacteria are often described as rhizosphere associated and depending on recently photosynthesized C sources, such as exudates (Denef et al. 2009; Mellado-Vázquez et al. 2016). In particular, the higher portions of plant-derived C in PLFA in the non-growing season points to a continued degradation of organic matter, though there is no input of fresh plant material. Furthermore, Denef et al. (2007) reported of a fungal-mediated translocation of rhizosphere-C from to bacterial biomass or a preferential bacterial use of fungal necromass several months after a pulse labelling. This mechanism of cross-feeding and recycling of carbon (Gleixner 2013) between fungi and G− bacteria might potentially explain the enrichment of markers assigned to G− bacteria in the non-growing season compared to the growing season, too. Therefore, we suggest that there was an increased saprotrophic assimilation of plant-derived C by G− bacteria. This is in line with recent findings (Bai et al. 2016), showing that both fungi and G− bacteria strongly rely on the decomposition of plant residues, when no recently photosynthesized C sources are available like in the non-growing season. Another possible explanation for our finding might be a fungal origin of the markers 16:1ω5 and 18:1ω9 which is discussed in several studies (Bååth and Anderson 2003; Kaiser et al. 2010; Madan et al. 2002; Olsson et al. 1995; Sakamoto et al. 2004). This suggests that the microbial origin of some PLFA markers depends strongly on substrate quality and availability (Borga et al. 1994; Madan et al. 2002; Wilkinson et al. 2002).
Conclusions
Our results demonstrate that the soil microbial community composition and their assimilation of plant-derived C can be independent of C3–C4 vegetation change. However, a selection of plants with similar characteristics is important so that the difference in photosynthetic pathways has little or no effect on the microbial community. Overall, our results suggest that C dynamics in soils strongly depend on C availability, which changes seasonally and favors different parts of the soil microbial community driving the soil C dynamics. On the other hand, soil texture was the most important factor influencing the soil microbial community composition in this study. Thus, contrary to the common idea that vegetation change experiments are biased by the preference of soil microorganisms to specific types of photosynthetic pathways, we show here that vegetation change experiments are robust to understand microbial C dynamics in soil. However, the results of this study are based on only a small number of plant species and two different soils. Therefore, future research should provide information on specific plant effects on the influence of C3–C4 vegetation changes on microbial soil communities. And secondly, the knowledge of how soil environmental parameters impact these effects, including the underlying mechanisms, should be expanded.
References
Bååth E, Anderson TH (2003) Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol Biochem 35(7):955–963
Bahn M, Lattanzi FA, Hasibeder R, Wild B, Koranda M, Danese V, Bruggemann N, Schmitt M, Siegwolf R, Richter A (2013) Responses of belowground carbon allocation dynamics to extended shading in mountain grassland. New Phytol 198(1):116–126
Bai Z, Liang C, Bode S, Huygens D, Boeckx P (2016) Phospholipid C-13 stable isotopic probing during decomposition of wheat residues. Appl Soil Ecol 98:65–74
Balesdent J, Balabane M (1996) Major contribution of roots to soil carbon storage inferred from maize cultivated soils. Soil Biol Biochem 28(9):1261–1263
Balser TC, Firestone MK (2005) Linking microbial community composition and soil processes in a California annual grassland and mixed-conifer forest. Biogeochemistry 73(2):395–415
Berg N, Steinberger Y (2010) Are biological effects of desert shrubs more important than physical effects on soil microorganisms? Microb Ecol 59(1):121–129
Black CA (1965) Methods of soil analysis: part I. Physical and mineralogical properties. American Society of Agronomy, Madison
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37(8):911–917
Borga P, Nilsson M, Tunlid A (1994) Bacterial communities in peat in relation to botanical composition as revealed by phospholipid fatty acid analysis. Soil Biol Biochem 26(7):841–848
Cao D, Shi F, Ruan W, Lu Z, Chai M (2011) Seasonal changes in and relationship between soil microbial and microfaunal communities in a Tamarix chinensis community in the Yellow River Delta. Afr J Biotechnol 10(80):18425–18432
Chen JR, Wang QL, Li M, Liu F, Li W (2016) Does the different photosynthetic pathway of plants affect soil respiration in a subtropical wetland? Ecol Evol 6(22):8010–8017
Cotrufo MF, Soong JL, Horton AJ, Campbell EE, Haddix ML, Wall DH, Parton AJ (2015) Formation of soil organic matter via biochemical and physical pathways of litter mass loss. Nat Geosci 8(10):776–779
Creamer CA, de Menezes AB, Krull ES, Sanderman J, Newton-Walters R, Farrell M (2015) Microbial community structure mediates response of soil C decomposition to litter addition and warming. Soil Biol Biochem 80:175–188
de Vries FT, Manning P, Tallowin JRB, Mortimer SR, Pilgrim ES, Harrison KA, Hobbs PJ, Quirk H, Shipley B, Cornelissen JHC, Kattge J, Bardgett RD (2012) Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities. Ecol Lett 15(11):1230–1239
Degens ET (1969) Biogeochemistry of stable carbon isotopes. In: Eglington GMMTJ (ed) Organic geochemistry. Elsevier, New York, pp 304–329
Denef K, Bubenheim H, Lenhart K, Vermeulen J, Van Cleemput O, Boeckx P, Müller C (2007) Community shifts and carbon translocation within metabolically-active rhizosphere microorganisms in grasslands under elevated CO2. Biogeosciences 4(5):769–779
Denef K, Roobroeck D, Manimel Wadu MCW, Lootens P, Boeckx P (2009) Microbial community composition and rhizodeposit-carbon assimilation in differently managed temperate grassland soils. Soil Biol Biochem 41(1):144–153
DeNiro M, Epstein S (1977) Mechanism of carbon isotope fractionation associated with lipid synthesis. Science 197(4300):261–263
Don A, Roedenbeck C, Gleixner G (2013) Unexpected control of soil carbon turnover by soil carbon concentration. Environ Chem Lett 11(4):407–413
Epron D, Ngao J, Dannoura M, Bakker MR, Zeller B, Bazot S, Bosc A, Plain C, Lata JC, Priault P, Barthes L, Loustau D (2011) Seasonal variations of belowground carbon transfer assessed by in situ 13CO2 pulse labelling of trees. Biogeosciences 8(5):1153–1168
Evershed RP, Crossman ZM, Bull ID, Mottram H, Dungait JAJ, Maxfield PJ, Brennand EL (2006) 13C-Labelling of lipids to investigate microbial communities in the environment. Curr Opin Biotechnol 17(1):72–82
Fierer N, Schimel JP, Holden PA (2003) Variations in microbial community composition through two soil depth profiles. Soil Biol Biochem 35(1):167–176
Frostegard A, Tunlid A, Baath E (1991) Microbial biomass measured as total lipid phosphate in soils of different organic content. J Microbiol Methods 14(3):151–163
Frostegard A, Tunlid A, Baath E (2011) Use and misuse of PLFA measurements in soils. Soil Biol Biochem 43(8):1621–1625
Garcia-Pausas J, Paterson E (2011) Microbial community abundance and structure are determinants of soil organic matter mineralisation in the presence of labile carbon. Soil Biol Biochem 43(8):1705–1713
Gleixner G (2013) Soil organic matter dynamics: a biological perspective derived from the use of compound-specific isotopes studies. Ecol Res 28(5):683–695
Gleixner G, Poirier N, Bol R, Balesdent J (2002) Molecular dynamics of organic matter in a cultivated soil. Org Geochem 33(3):357–366
Grayston SJ, Griffith GS, Mawdsley JL, Campbell CD, Bardgett RD (2001) Accounting for variability in soil microbial communities of temperate upland grassland ecosystems. Soil Biol Biochem 33(4–5):533–551
Habekost M, Eisenhauer N, Scheu S, Steinbeiss S, Weigelt A, Gleixner G (2008) Seasonal changes in the soil microbial community in a grassland plant diversity gradient four years after establishment. Soil Biol Biochem 40(10):2588–2595
Hungate BA, Mau RL, Schwartz E, Caporaso JG, Dijkstra P, van Gestel N, Koch BJ, Liu CM, McHugh TA, Marks JC, Morrissey EM, Price LB (2015) Quantitative microbial ecology through stable isotope probing. Appl Environ Microbiol 81(21):7570–7581
Joergensen RG, Wichern F (2008) Quantitative assessment of the fungal contribution to microbial tissue in soil. Soil Biol Biochem 40(12):2977–2991
Johnson MJ, Lee KY, Scow KM (2003) DNA fingerprinting reveals links among agricultural crops, soil properties, and the composition of soil microbial communities. Geoderma 114(3–4):279–303
Kaiser C, Frank A, Wild B, Koranda M, Richter A (2010) Negligible contribution from roots to soil-borne phospholipid fatty acid fungal biomarkers 18:2ω6,9 and 18:1ω9. Soil Biol Biochem 42(9):1650–1652
Kaur A, Chaudhary A, Choudhary R, Kaushik R (2005) Phospholipid fatty acid—a bioindicator of environment monitoring and assessment in soil ecosystem. Curr Sci 89(7):1103–1112
Kindler R, Miltner A, Thullner M, Richnow HH, Kästner M (2009) Fate of bacterial biomass derived fatty acids in soil and their contribution to soil organic matter. Org Geochem 40(1):29–37
Klamer M, Hedlund K (2004) Fungal diversity in set-aide agricultural soil investigated using terminal-restriction fragment length polymorphism. Soil Biol Biochem 36(6):983–988
Kramer C, Gleixner G (2006) Variable use of plant- and soil-derived carbon by microorganisms in agricultural soils. Soil Biol Biochem 38(11):3267–3278
Kroppenstedt RM (1985) Fatty acid and menaquinone analysis of actinomycetes and related organisms. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 173–199
Lal R (2004) Soil carbon sequestration impacts on global climate change and food security. Science 304(5677):1623–1627
Lange M, Habekost M, Eisenhauer N, Roscher C, Bessler H, Engels C, Oelmann Y, Scheu S, Wilcke W, Schulze ED, Gleixner G (2014) Biotic and abiotic properties mediating plant diversity effects on soil microbial communities in an experimental grassland. PLoS ONE 9(5):e96182
Lange M, Eisenhauer N, Sierra CA, Bessler H, Engels C, Griffiths RI, Mellado-Vazquez PG, Malik AA, Roy J, Scheu S, Steinbeiss S, Thomson BC, Trumbore SE, Gleixner G (2015) Plant diversity increases soil microbial activity and soil carbon storage. Nat Commun 6:6707
Liang C, Schimel JP, Jastrow JD (2017) The importance of anabolism in microbial control over soil carbon storage. Nat Microbiol 2(8):17105
Madan R, Pankhurst C, Hawke B, Smith S (2002) Use of fatty acids for identification of AM fungi and estimation of the biomass of AM spores in soil. Soil Biol Biochem 34(1):125–128
Malik A, Scheibe A, LokaBharathi PA, Gleixner G (2012) Online stable isotope analysis of dissolved organic carbon size classes using size exclusion chromatography coupled to an isotope ratio mass spectrometer. Environ Sci Technol 46(18):10123–10129
Mau RL, Liu CM, Aziz M, Schwartz E, Dijkstra P, Marks JC, Price LB, Keim P, Hungate BA (2015) Linking soil bacterial biodiversity and soil carbon stability. ISME J 9(6):1477–1480
Medeiros PM, Fernandes MF, Dick RP, Simoneit BRT (2006) Seasonal variations in sugar contents and microbial community in a ryegrass soil. Chemosphere 65(5):832–839
Mellado-Vázquez PG, Lange M, Bachmann D, Gockele A, Karlowsky S, Milcu A, Piel C, Roscher C, Roy J, Gleixner G (2016) Plant diversity generates enhanced soil microbial access to recently photosynthesized carbon in the rhizosphere. Soil Biol Biochem 94:122–132
Merckx R, den Hartog A, van Veen JA (1985) Turnover of root-derived material and related microbial biomass formation in soils of different texture. Soil Biol Biochem 17(4):565–569
Oksanen J, Blanchet FG, Kindt R, Legendre P, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2011) vegan: Community Ecology Package. R package version 1.17-9
O’Leary MH (1981) Carbon isotope fractionation in plants. Phytochemistry 20(4):553–567
Olsson PA, Baath E, Jakobsen I, Soderstrom B (1995) The use of phospholipid and neutral lipid fatty acids to estimate biomass of arbuscular mycorrhizal fungi in soil. Mycol Res 99(5):623–629
Pett-Ridge J, Firestone MK (2017) Using stable isotopes to explore root-microbe-mineral interactions in soil. Rhizosphere 3(Part 2):244–253
Porazinska DL, Bardgett RD, Blaauw MB, Hunt H, William HW, Parsons AN, Seastedt TR, Wall DH (2003) Relationships at the aboveground-belowground interface: plants, soil biota, and soil processes. Ecol Monogr 73(3):377–395
Sakamoto K, Iijima T, Higuchi R (2004) Use of specific phospholipid fatty acids for identifying and quantifying the external hyphae of the arbuscular mycorrhizal fungus Gigaspora rosea. Soil Biol Biochem 36(11):1827–1834
Scheibe A, Krantz L, Gleixner G (2012) Simultaneous determination of the quantity and isotopic signature of dissolved organic matter from soil water using high-performance liquid chromatography/isotope ratio mass spectrometry. Rapid Commun Mass Spectrom 26(2):173–180
Singh BK, Munro S, Reid E, Ord B, Potts JM, Paterson E, Millard P (2006) Investigating microbial community structure in soils by physiological, biochemical and molecular fingerprinting methods. Eur J Soil Sci 57(1):72–82
Steinbeiss S, Temperton VM, Gleixner G (2008) Mechanisms of short-term soil carbon storage in experimental grasslands. Soil Biol Biochem 40(10):2634–2642
ter Braak CJF, Šmilauer P (2012) Canoco reference manual and user’s guide: software for ordination (version 5.0). Microcomputer Power, Ithaca
Thoms C, Gleixner G (2013) Seasonal differences in tree species’ influence on soil microbial communities. Soil Biol Biochem 66:239–248
Thoms C, Gattinger A, Jacob M, Thomas FM, Gleixner G (2010) Direct and indirect effects of tree diversity drive soil microbial diversity in temperate deciduous forest. Soil Biol Biochem 42(9):1558–1565
Treonis AM, Ostle NJ, Stott AW, Primrose R, Grayston SJ, Ineson P (2004) Identification of groups of metabolically-active rhizosphere microorganisms by stable isotope probing of PLFAs. Soil Biol Biochem 36(3):533–537
Voroney RP, Heck RJ (2015) The soil habitat. In: Paul EA (ed) Soil microbiology, ecology and biochemistry, 4th edn. Academic Press, Boston, pp 15–39
Wardle DA (2002) Linking the aboveground and belowground components. In: Levin SA, Horn HS (eds) Monographs in population biology. Princeton University Press, Princeton
Werner RA, Brand WA (2001) Referencing strategies and techniques in stable isotope ratio analysis. Rapid Commun Mass Spectrom 15(7):501–519
Wilkinson SC, Anderson JM, Scardelis SP, Tisiafouli M, Taylor A, Wolters V (2002) PLFA profiles of microbial communities in decomposing conifer litters subject to moisture stress. Soil Biol Biochem 34(2):189–200
Zelles L (1997) Phospholipid fatty acid profiles in selected members of soil microbial communities. Chemosphere 35(1–2):275–294
Zelles L (1999) Identification of single cultured micro-orgnisms based on their whole-community fatty acid profiles, using an extended extraction procedure. Chemosphere 39(4):665–682
Zuur AF, Ieno EN, Walker N, Saveliev AA, Smith GM (2009) Mixed effects models and extensions in ecology with R. Springer New York, New York
Acknowledgements
Open access funding provided by Max Planck Society. The authors would like to thank Uta Gerighausen and Ernesto Rendón for their help during soil sampling, Agnes Fastnacht for maintaining and weeding the experimental site, and Steffen Rühlow for assistance with the GC-FID and GC-IRMS systems. We would also like to thank three anonymous reviewers and associate editor Susan Ziegler for their reviews that helped to improve the manuscript. The International Max Planck Research School (IMPRS) of the Max Planck Institute for Biogeochemistry provided funding for the PhD scholarship of Perla G. Mellado-Vázquez. Markus Lange is supported by the Max Planck Institute for Biogeochemistry, Jena, Germany and is funded by the German Research Foundation (DFG; FOR 456, FOR 1451 – “The Jena Experiment”) and by the “Zwillenberg-Tietz Stiftung”. We also appreciate the support of the Friedrich Schiller University of Jena.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Susan Ziegler.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Mellado-Vázquez, P.G., Lange, M. & Gleixner, G. Soil microbial communities and their carbon assimilation are affected by soil properties and season but not by plants differing in their photosynthetic pathways (C3 vs. C4). Biogeochemistry 142, 175–187 (2019). https://doi.org/10.1007/s10533-018-0528-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10533-018-0528-9