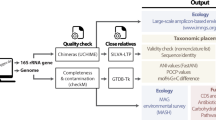
Abstract
Bacterial species are commonly defined by applying a set of predetermined criteria, including DNA–DNA hybridization values, 16S rRNA gene sequence similarity, phenotypic data as well as genome-based criteria such as average nucleotide identity or digital DNA-DNA hybridization. These criteria mostly allow for the delimitation of taxa that resemble typical bacterial species. Their application is often complicated when the objective is to delineate new species that are characterized by significant population-level diversity or recent speciation. However, we believe that these complexities and limitations can be easily circumvented by recognizing that bacterial species represent unique and exclusive assemblages of diversity. Within such a framework, methods that account for the population processes involved in species evolution are used to infer species boundaries. A method such as genealogical concordance analysis is well suited to delineate a putative species. The existence of the new taxon is then interrogated using an array of traditional and genome-based characters. By making use of taxa in the genera Pantoea, Paraburkholderia and Escherichia we demonstrate in a step-wise process how genealogical concordance can be used to delimit a bacterial species. Genetic, phenotypic and biological criteria were used to provide independent lines of evidence for the existence of that taxon. Our six-step approach to species recognition is straightforward and applicable to bacterial species especially in the post-genomic era, with increased availability of whole genome sequences. In fact, our results indicated that a combined genome-based comparative and evolutionary approach would be the preferred alternative for delineating coherent bacterial taxa.





Similar content being viewed by others
References
Achtman M, Wagner M (2008) Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol 6:431–440
Avise JC, Ball RM (1990) Principles of genealogical concordance in species concepts and biological taxonomy. Oxf Surv Evol Biol 7:23–45
Baltrus DA, Mccann HC, Guttman DS (2017) Evolution, genomics and epidemiology of Pseudomonas syringae: challenges in bacterial molecular plant pathology. Mol Plant Pathol 18:152–168
Baum DA (2007) Concordance trees, concordance factors, and the exploration of reticulate genealogy. Taxon 56:417–426
Baum DA, Shaw KL (1995) Genealogical perspectives on the species problem. In: Hoch PC, Stephenson AG (eds) Experimental and molecular approaches to plant biosystematics. Missouri Botanical Garden, St Louis, pp 289–303
Berthe T, Ratajczak M, Clermont O, Denamur E, Petit F (2013) Evidence for coexistence of distinct Escherichia coli populations in various aquatic environments and their survival in estuary water. Appl Environ Microbiol 79:4684–4693
Beukes CW, Venter SN, Law IJ, Phalane FL, Steenkamp ET (2013) South African Papilionoid legumes are nodulated by diverse Burkholderia with unique nodulation and nitrogen-fixation loci. PLoS ONE 8(7):e68406
Booth A, Mariscal C, Doolittle WF (2016) The modern synthesis in the light of microbial genomics. Annu Rev Microbiol 70:279–297
Boucher Y, Nesbo CL, Doolittle WF (2001) Microbial genomes: dealing with diversity. Curr Opin Microbiol 4:285–289
Brady C, Cleenwerck I, Venter S, Vancanneyt M, Swings J, Coutinho T (2008) Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Syst Appl Microbiol 31:447–460
Brady CL, Goszczynska T, Venter SN, Cleenwerck I, De Vos P, Gitaitis RD, Coutinho TA (2011) Pantoea allii sp. nov., isolated from onion plants and seed. Int J Syst Evol Microbiol 61:932–937
Brady C, Cleenwerck I, Venter S, Coutinho T, De Vos P (2013) Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov as Lelliottia nimipressuralis comb. nov and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov as Pluralibacter gergoviae comb. nov and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst Appl Microbiol 36:309–319
Brisse S, Passet V, PaD Grimont (2014) Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int J Syst Evol Microbiol 64:3146–3152
Clermont O, Gordon DM, Brisse S, Walk ST, Denamur E (2011) Characterization of the cryptic Escherichia lineages: rapid identification and prevalence. Environ Microbiol 13:2468–2477
Colwell RR (1970) Polyphasic taxonomy of the genus Vibrio: numerical taxonomy of Vibrio cholerae, Vibrio parahaemolyticus, and related Vibrio species. J Bacteriol 104:410–433
Coutinho T, Venter S (2009) Pantoea ananatis: an unconventional plant pathogen. Mol Plant Pathol 10:325–335
Coyne J, Orr H (2004) Speciation. Sinauer Associates, Sunderland
Dayrat B (2005) Towards integrative taxonomy. Biol J Linn Soc 85:407–415
De Queiroz K (2005) Ernst Mayr and the modern concept of species. Proc Natl Acad Sci USA 102(Suppl 1):6600–6607
De Queiroz A, Donoghue MJ, Kim J (1995) Separate versus combined analysis of phylogenetic evidence. Annu Rev Ecol Syst 26:657–681
Dobritsa AP, Samadpour M (2016) Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int J Syst Evol Microbiol 66:2836–2846
Dykhuizen DE, Green L (1991) Recombination in Escherichia coli and the definition of biological species. J Bacteriol 173:7257–7268
Ereshefsky M (2011) Mystery of mysteries: darwin and the species problem. Cladistics 27:67–79
Felsenstein J (2004) Inferring phylogenies. Sinauer Associates, Sunderland
Galtier N, Daubin V (2008) Dealing with incongruence in phylogenomic analyses. Philos Trans R Soc Lond B Biol Sci 363:4023–4029
Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, Stackebrandt E, De Peer YV, Vandamme P, Thompson FL, Swings J (2005) Re-evaluating prokaryotic species. Nat Rev Microbiol 3:733–739
Ghai R, Martin-Cuadrado A-B, Molto AG, Heredia IG, Cabrera R, Martin J, Verdu M, Deschamps P, Moreira D, Lopez-Garcia P, Mira A, Rodriguez-Valera F (2010) Metagenome of the Mediterranean deep chlorophyll maximum studied by direct and fosmid library 454 pyrosequencing. ISME J 4:1154–1166
Glaeser SP, Kampfer P (2015) Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst Appl Microbiol 38:237–245
Gontcharov AA, Marin B, Melkonian M (2004) Are combined analyses better than single gene phylogenies? A case study using SSU rDNA and rbcL sequence comparisons in the Zygnematophyceae (Streptophyta). Mol Biol Evol 21:612–624
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91
Hennig W (1999) Phylogenetic systematics. University of Illinois Press, Urbana
Hey J (2001) The mind of the species problem. Trends Ecol Evol 16:326–329
Holder M, Lewis PO (2003) Phylogeny estimation: traditional and Bayesian approaches. Nat Rev Genet 4:275–284
Jain R, Rivera MC, Lake JA (1999) Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci USA 96:3801–3806
Konstantinidis KT, Tiedje JM (2005) Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA 102:2567–2572
Leslie JF, Zeller KA, Summerell BA (2001) Icebergs and species in populations of Fusarium. Physiol Mol Plant Path 59:107–117
Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT (2011) Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci USA 108:7200–7205
Maddison WP (1997) Gene trees in species trees. Syst Biol 46:523–536
Mallet J (2001) Species, concepts of. In: Levin SA (ed) Encyclopedia of biodiversity, vol 5. Academic Press, New York, pp 427–440
Mergaert J, Verdonck L, Kersters K (1993) Transfer of Erwinia ananas (synonym, Erwinia uredovora) and Erwinia stewartii to the genus Pantoea emend. as Pantoea ananas (Serrano 1928) comb. nov. and Pantoea stewartii (Smith 1898) comb. nov., respectively, and description of Pantoea stewartii subsp. indologenes subsp. nov. Int J Syst Bacteriol 43:162–173
Naser SM, Dawyndt P, Hoste B, Gevers D, Vandemeulebroecke K, Cleenwerck I, Vancanneyt M, Swings J (2007) Identification of lactobacilli by pheS and rpoA gene sequence analyses. Int J Syst Evol Microbiol 57:2777–2789
Ochman H, Lawrence JG, Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304
Padial JM, Miralles A, De La Riva I, Vences M (2010) The integrative future of taxonomy. Front Zool 7:16
Philippe H, Douady CJ (2003) Horizontal gene transfer and phylogenetics. Curr Opin Microbiol 6:498–505
Retchless AC, Lawrence JG (2007) Temporal fragmentation of speciation in bacteria. Science 317:1093–1096
Richter M, Rossello-Mora R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Rising JD, Avise JC (1993) Application of genealogical-concordance principles to the taxonomy and evolutionary history of the sharp-tailed sparrow (Ammodramus caudacutus). Auk 110:844–856
Rokas A, Williams BL, King N, Carroll SB (2003) Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425:798–804
Rossello-Mora R, Amann R (2001) The species concept for prokaryotes. FEMS Microbiol Rev 25:39–67
Rossello-Mora R, Amann R (2015) Past and future species definitions for bacteria and archaea. Syst Appl Microbiol 38:209–216
Rosselló-Mora R, López-López A (2008) The least common denominator: Species or operational taxonomic units? In: Zengler K (ed) Accessing uncultivated microorganisms. American Society for Microbiology, Washington, DC, pp 117–130
Salichos L, Rokas A (2013) Inferring ancient divergences requires genes with strong phylogenetic signals. Nature 497:327–331
Sites JW, Marshall JC (2004) Operational criteria for delimiting species. Annu Rev Ecol Evol Syst 35:199–227
Stackebrandt E, Frederiksen W, Garrity GM, Grimont PA, Kampfer P, Maiden MC, Nesme X, Rossello-Mora R, Swings J, Truper HG, Vauterin L, Ward AC, Whitman WB (2002) Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol 52:1043–1047
Staley JT (2006) The bacterial species dilemma and the genomic-phylogenetic species concept. Philos Trans R Soc Lond B Biol Sci 361:1899–1909
Steenkamp ET, Van Zyl E, Beukes CW, Avontuur JR, Chan WY, Palmer M, Mthombeni LS, Phalane FL, Sereme TK, Venter SN (2015) Burkholderia kirstenboschensis sp. nov. nodulates papilionoid legumes indigenous to South Africa. Syst Appl Microbiol 38:545–554
Sutcliffe IC (2015) Challenging the anthropocentric emphasis on phenotypic testing in prokaryotic species descriptions: rip it up and start again. Front Genet 6:218
Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32
Thiergart T, Landan G, Martin WF (2014) Concatenated alignments and the case of the disappearing tree. BMC Evol Biol 14:266
Thompson CC, Chimetto L, Edwards RA, Swings J, Stackebrandt E, Thompson FL (2013) Microbial genomic taxonomy. BMC Genom 14:913
Tibayrenc M (1999) Toward an integrated genetic epidemiology of parasitic protozoa and other pathogens. Annu Rev Genet 33:449–477
Tindall BJ, Rossello-Mora R, Busse HJ, Ludwig W, Kampfer P (2010) Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 60:249–266
Vandamme P, Peeters C (2014) Time to revisit polyphasic taxonomy. Antonie Van Leeuwenhoek 106:57–65
Vandamme P, Pot B, Gillis M, De Vos P, Kersters K, Swings J (1996) Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev 60:407–438
Vandamme P, Moore ER, Cnockaert M, De Brandt E, Svensson-Stadler L, Houf K, Spilker T, Lipuma JJ (2013) Achromobacter animicus sp. nov., Achromobacter mucicolens sp. nov., Achromobacter pulmonis sp. nov. and Achromobacter spiritinus sp. nov., from human clinical samples. Syst Appl Microbiol 36:1–10
Vanlaere E, Baldwin A, Gevers D, Henry D, De Brandt E, Lipuma JJ, Mahenthiralingam E, Speert DP, Dowson C, Vandamme P (2009) Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int J Syst Evol Microbiol 59:102–111
Walk ST, Alm EW, Gordon DM, Ram JL, Toranzos GA, Tiedje JM, Whittam TS (2009) Cryptic lineages of the genus Escherichia. Appl Environ Microbiol 75:6534–6544
Wayne LG, Brenner DJ, Colwell RR, PaD Grimont, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Truper HG (1987) Report of the Ad Hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol 37:463–464
Wilson EO, Brown WL (1953) The subspecies concept and its taxonomic application. Syst Zool 2:97–111
Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M (2006) Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151
Acknowledgements
We are grateful to the South African National Research Foundation (NRF) and the Department of Science and Technology for funding through their Center of Excellence programme. Dr. Seth Walk from the Department of Microbiology and Immunology, Montana State University for sharing the original E. coli MLST dataset published in Walk et al. (2009).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Venter, S.N., Palmer, M., Beukes, C.W. et al. Practically delineating bacterial species with genealogical concordance. Antonie van Leeuwenhoek 110, 1311–1325 (2017). https://doi.org/10.1007/s10482-017-0869-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-017-0869-8