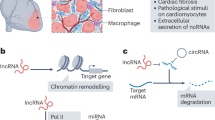
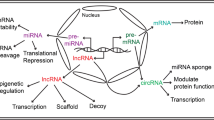
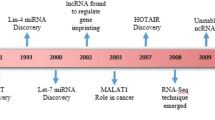
Abstract
Cardiovascular diseases (CVDs) are the leading causes of death worldwide. Increasing reports demonstrated that non-coding RNAs (ncRNAs) have been crucially involved in the development of CVDs. Piwi-interacting RNAs (piRNAs) are a novel cluster of small non-coding RNAs with strong uracil bias at the 5′ end and 2′-O-methylation at the 3′ end that are mainly present in the mammalian reproductive system and stem cells and serve as potential modulators of developmental and pathophysiological processes. Recently, piRNAs have been reported to be widely expressed in human tissues and can potentially regulate various diseases. Specifically, concomitant with the development of next-generation sequencing techniques, piRNAs have been found to be differentially expressed in CVDs, indicating their potential involvement in the occurrence and progression of heart diseases. However, the molecular mechanisms and signaling pathways involved with piRNA function have not been fully elucidated. In this review, we present the current understanding of the piRNAs from the perspectives of biogenesis, characteristics, biological function, and regulatory mechanisms, and highlight their potential roles and underlying mechanisms in CVDs, which will provide new insights into the potential applications of piRNAs in the clinical diagnosis, prognosis, and therapeutic strategies for heart diseases.




Similar content being viewed by others
Abbreviations
- ADSCs:
-
Adipose-derived stem cells
- AGO2:
-
Argonaute 2
- ARMI:
-
Armitage
- Aub:
-
Aubergine
- Ccr4-NOT:
-
Glucose-repressible alcohol dehydrogenase transcriptional effector
- DNMT:
-
DNA methyltransferase
- CVDs:
-
Cardiovascular diseases
- Cdk5rap1:
-
CDK5 regulatory subunit-associated protein 1
- CRISPR-Cas:
-
Clustered regularly interspaced short palindromic repeats/CRISPR-associated proteins
- CTEPH:
-
Chronic thromboembolic pulmonary hypertension
- dsRNA:
-
Double-stranded RNA
- ECs:
-
Endothelial cells
- HF:
-
Heart failure
- H3K9me3:
-
Histone 3 trimethylated on lysine 9
- LINE-1:
-
Long interspersed nuclear elements-1
- MI:
-
Myocardial infarction
- MCAF1:
-
Activating transcription factor 7 interacting protein
- MECP2:
-
Methyl CpG binding protein 2
- MM:
-
Multiple myeloma
- MOV10L1:
-
Mov10-like RISC complex RNA helicase 1
- MSCs:
-
Mesenchymal stem cells
- ncRNAs:
-
Non-coding RNAs
- Nbr:
-
Nibbler
- PH:
-
Pulmonary hypertension
- piRNAs:
-
Piwi-interacting RNAs
- piRISCs:
-
PiRNA-induced silence compounds
- PRMT5:
-
The type II arginine methyltransferasev
- qPCR:
-
Quantitative PCR
- RNAi:
-
RNA interference
- RNA-seq:
-
RNA sequencing
- SETDB1:
-
SET domain-bifurcated histone lysine methyltransferase 1
- siRNA:
-
Small interfering RNA
- SUMO:
-
Small ubiquitin-like modifier
- TE:
-
Transposable element
- UDRs:
-
UORF downstream regions
- UHRF1:
-
Ubiquitin-like, containing PHD and RING finger domains 1
- uORFs:
-
Proximal short open reading frames
- WDR77:
-
WD repeat domain 77
- Zuc:
-
Zucchini
References
Shi Q, Yang X (2016) Circulating MicroRNA and long noncoding RNA as biomarkers of cardiovascular diseases. J Cell Physiol 231(4):751–755. https://doi.org/10.1002/jcp.25174
Liu Y, Yang Y, Wang Z, Fu X, Chu X-M, Li Y, Wang Q, He X, Li M, Wang K, Wang J-X, Li P-F, Yu T (2020) Insights into the regulatory role of circRNA in angiogenesis and clinical implications. Atherosclerosis 298:14–26. https://doi.org/10.1016/j.atherosclerosis.2020.02.017
Liu S, Yang Y, Jiang S, Xu H, Tang N, Lobo A, Zhang R, Liu S, Yu T, Xin H (2019) MiR-378a-5p regulates proliferation and migration in vascular smooth muscle cell by targeting CDK1. Front Genet 10:22. https://doi.org/10.3389/fgene.2019.00022
Tang N, Jiang S, Yang Y, Liu S, Ponnusamy M, Xin H, Yu T (2018) Noncoding RNAs as therapeutic targets in atherosclerosis with diabetes mellitus. Cardiovasc Ther 36(4):e12436. https://doi.org/10.1111/1755-5922.12436
Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li X, Yang W, Zhang C, Yang Q, Yee A, Chen Y, Yang F, Sun H, Huang R, Yee AJ, Li R-K, Wu Z, Backx PH, Yang BB (2017) A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics 7(16):3842–3855. https://doi.org/10.7150/thno.19764
Sharma AK, Nelson MC, Brandt JE, Wessman M, Mahmud N, Weller KP, Hoffman R (2001) Human CD34(+) stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood 97(2):426–434
Girard A, Sachidanandam R, Hannon GJ, Carmell MA (2006) A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442(7099):199–202. https://doi.org/10.1038/nature04917
Perera BPU, Tsai ZT-Y, Colwell ML, Jones TR, Goodrich JM, Wang K, Sartor MA, Faulk C, Dolinoy DC (2019) Somatic expression of piRNA and associated machinery in the mouse identifies short, tissue-specific piRNA. Epigenetics 14(5):504–521. https://doi.org/10.1080/15592294.2019.1600389
Roy J, Sarkar A, Parida S, Ghosh Z, Mallick B (2017) Small RNA sequencing revealed dysregulated piRNAs in Alzheimer's disease and their probable role in pathogenesis. Mol Biosyst 13(3):565–576. https://doi.org/10.1039/c6mb00699j
Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath BS, Kosik KS (2011) Identification of piRNAs in the central nervous system. RNA 17(6):1090–1099. https://doi.org/10.1261/rna.2565011
Moyano M, Stefani G (2015) piRNA involvement in genome stability and human cancer. J Hematol Oncol 8:38. https://doi.org/10.1186/s13045-015-0133-5
Rajan KS, Velmurugan G, Pandi G, Ramasamy S (2014) miRNA and piRNA mediated Akt pathway in heart: antisense expands to survive. Int J Biochem Cell Biol 55:153–156. https://doi.org/10.1016/j.biocel.2014.09.001
Lipps C, Northe P, Figueiredo R, Rohde M, Brahmer A, Krämer-Albers E-M, Liebetrau C, Wiedenroth CB, Mayer E, Kriechbaum SD, Dörr O, Nef H, Hamm CW, Keller T, Troidl C (2019) Non-invasive approach for evaluation of pulmonary hypertension using extracellular vesicle-associated small non-coding RNA. Biomolecules. https://doi.org/10.3390/biom9110666
Rajan KS, Velmurugan G, Gopal P, Ramprasath T, Babu DDV, Krithika S, Jenifer YC, Freddy A, William G, Kalpana K, Ramasamy S (2016) Abundant and altered expression of PIWI-interacting RNAs during cardiac hypertrophy. Heart Lung Circ 25(10):1013–1020. https://doi.org/10.1016/j.hlc.2016.02.015
Vella S, Gallo A, Lo Nigro A, Galvagno D, Raffa GM, Pilato M, Conaldi PG (2016) PIWI-interacting RNA (piRNA) signatures in human cardiac progenitor cells. Int J Biochem Cell Biol. https://doi.org/10.1016/j.biocel.2016.04.012
Li Y, Zeng A, Li G, Guan Y-N, Yang H-T, Shen B, Jing Q (2017) Dynamic regulation of small RNAome during the early stage of cardiac differentiation from pluripotent embryonic stem cells. Genom Data 12:136–145. https://doi.org/10.1016/j.gdata.2017.05.006
Yang J, Xue FT, Li YY, Liu W, Zhang S (2018) Exosomal piRNA sequencing reveals differences between heart failure and healthy patients. Eur Rev Med Pharmacol Sci 22(22):7952–7961. https://doi.org/10.26355/eurrev_201811_16423
Izumi N, Shoji K, Suzuki Y, Katsuma S, Tomari Y (2020) Zucchini consensus motifs determine the mechanism of pre-piRNA production. Nature 578(7794):311–316. https://doi.org/10.1038/s41586-020-1966-9
Cheng Y, Wang Q, Jiang W, Bian Y, Zhou Y, Gou A, Zhang W, Fu K, Shi W (2019) Emerging roles of piRNAs in cancer: challenges and prospects. Aging (Albany NY) 11(21):9932–9946. https://doi.org/10.18632/aging.102417
Rajan KS, Ramasamy S (2014) Retrotransposons and piRNA: the missing link in central nervous system. Neurochem Int. https://doi.org/10.1016/j.neuint.2014.05.017
Mao Y, Qian S-B (2020) Ribosome-guided piRNA production. Nat Cell Biol 22(2):141–142. https://doi.org/10.1038/s41556-020-0464-5
Sun YH, Zhu J, Xie LH, Li Z, Meduri R, Zhu X, Song C, Chen C, Ricci EP, Weng Z, Li XZ (2020) Ribosomes guide pachytene piRNA formation on long intergenic piRNA precursors. Nat Cell Biol 22(2):200–212. https://doi.org/10.1038/s41556-019-0457-4
Iwasaki YW, Siomi MC, Siomi H (2015) PIWI-interacting RNA: its biogenesis and functions. Annu Rev Biochem 84:405–433. https://doi.org/10.1146/annurev-biochem-060614-034258
Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RHA, Hannon GJ, Draper BW, Ketting RF (2007) A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129(1):69–82
Freedman JE, Gerstein M, Mick E, Rozowsky J, Levy D, Kitchen R, Das S, Shah R, Danielson K, Beaulieu L, Navarro FCP, Wang Y, Galeev TR, Holman A, Kwong RY, Murthy V, Tanriverdi SE, Koupenova-Zamor M, Mikhalev E, Tanriverdi K (2016) Diverse human extracellular RNAs are widely detected in human plasma. Nat Commun 7:11106. https://doi.org/10.1038/ncomms11106
Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T (2007) The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol 14(4):349–350
Yamaguchi S, Oe A, Nishida KM, Yamashita K, Kajiya A, Hirano S, Matsumoto N, Dohmae N, Ishitani R, Saito K, Siomi H, Nishimasu H, Siomi MC, Nureki O (2020) Crystal structure of Drosophila Piwi. Nat Commun 11(1):858. https://doi.org/10.1038/s41467-020-14687-1
Gainetdinov I, Colpan C, Arif A, Cecchini K, Zamore PD (2018) A single mechanism of biogenesis, initiated and directed by PIWI proteins, explains piRNA production in most animals. Mol Cell. https://doi.org/10.1016/j.molcel.2018.08.007
Nagamori I, Kobayashi H, Nishimura T, Yamagishi R, Katahira J, Kuramochi-Miyagawa S, Kono T, Nakano T (2018) Relationship between PIWIL4-mediated H3K4me2 demethylation and piRNA-dependent DNA methylation. Cell Rep 25(2):350–356. https://doi.org/10.1016/j.celrep.2018.09.017
Schulze M, Sommer A, Plötz S, Farrell M, Winner B, Grosch J, Winkler J, Riemenschneider MJ (2018) Sporadic Parkinson's disease derived neuronal cells show disease-specific mRNA and small RNA signatures with abundant deregulation of piRNAs. Acta Neuropathol Commun 6(1):58. https://doi.org/10.1186/s40478-018-0561-x
Qiu W, Guo X, Lin X, Yang Q, Zhang W, Zhang Y, Zuo L, Zhu Y, Li C-SR, Ma C, Luo X (2017) Transcriptome-wide piRNA profiling in human brains of Alzheimer's disease. Neurobiol Aging 57:170–177. https://doi.org/10.1016/j.neurobiolaging.2017.05.020
Guo B, Li D, Du L, Zhu X (2020) piRNAs: biogenesis and their potential roles in cancer. Cancer Metastasis Rev. https://doi.org/10.1007/s10555-020-09863-0
Das B, Jain N, Mallick B (2020) piR-39980 promotes cell proliferation, migration and invasion, and inhibits apoptosis via repression of SERPINB1 in human osteosarcoma. Biol Cell 112(3):73–91. https://doi.org/10.1111/boc.201900063
Zhang L, Meng X, Pan C, Qu F, Gan W, Xiang Z, Han X, Li D (2020) piR-31470 epigenetically suppresses the expression of glutathione S-transferase pi 1 in prostate cancer via DNA methylation. Cell Signal 67:109501. https://doi.org/10.1016/j.cellsig.2019.109501
Manakov SA, Pezic D, Marinov GK, Pastor WA, Sachidanandam R, Aravin AA (2015) MIWI2 and MILI have differential effects on piRNA biogenesis and DNA methylation. Cell Rep 12(8):1234–1243. https://doi.org/10.1016/j.celrep.2015.07.036
Kojima-Kita K, Kuramochi-Miyagawa S, Nagamori I, Ogonuki N, Ogura A, Hasuwa H, Akazawa T, Inoue N, Nakano T (2016) MIWI2 as an effector of DNA methylation and gene silencing in embryonic male germ cells. Cell Rep 16(11):2819–2828. https://doi.org/10.1016/j.celrep.2016.08.027
Watanabe T, Cui X, Yuan Z, Qi H, Lin H (2018) MIWI2 targets RNAs transcribed from piRNA-dependent regions to drive DNA methylation in mouse prospermatogonia. EMBO J. https://doi.org/10.15252/embj.201695329
Zhang H, Gao Q, Tan S, You J, Lyu C, Zhang Y, Han M, Chen Z, Li J, Wang H, Liao L, Qin J, Li J, Wong J (2019) SET8 prevents excessive DNA methylation by methylation-mediated degradation of UHRF1 and DNMT1. Nucleic Acids Res 47(17):9053–9068. https://doi.org/10.1093/nar/gkz626
Dong J, Wang X, Cao C, Wen Y, Sakashita A, Chen S, Zhang J, Zhang Y, Zhou L, Luo M, Liu M, Liao A, Namekawa SH, Yuan S (2019) UHRF1 suppresses retrotransposons and cooperates with PRMT5 and PIWI proteins in male germ cells. Nat Commun 10(1):4705. https://doi.org/10.1038/s41467-019-12455-4
Vaughan RM, Rothbart SB, Dickson BM (2019) The finger loop of the SRA domain in the E3 ligase UHRF1 is a regulator of ubiquitin targeting and is required for the maintenance of DNA methylation. J Biol Chem 294(43):15724–15732. https://doi.org/10.1074/jbc.RA119.010160
Su J-F, Zhao F, Gao Z-W, Hou Y-J, Li Y-Y, Duan L-J, Lun S-M, Yang H-J, Li J-K, Dai N-T, Shen F-F, Zhou F-Y (2020) piR-823 demonstrates tumor oncogenic activity in esophageal squamous cell carcinoma through DNA methylation induction via DNA methyltransferase 3B. Pathol Res Pract. https://doi.org/10.1016/j.prp.2020.152848
Zhang C, Sha H, Peng Y, Wang Y, Liu C, Zhou X (2019) PiRNA-DQ541777 contributes to neuropathic pain via targeting Cdk5rap1. J Neurosci 39(45):9028–9039. https://doi.org/10.1523/JNEUROSCI.1602-19.2019
Holoch D, Moazed D (2015) RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16(2):71–84. https://doi.org/10.1038/nrg3863
Sienski G, Batki J, Senti K-A, Dönertas D, Tirian L, Meixner K, Brennecke J (2015) Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery. Genes Dev 29(21):2258–2271. https://doi.org/10.1101/gad.271908.115
Yu Y, Gu J, Jin Y, Luo Y, Preall JB, Ma J, Czech B, Hannon GJ (2015) Panoramix enforces piRNA-dependent cotranscriptional silencing. Science 350(6258):339–342. https://doi.org/10.1126/science.aab0700
Ninova M, Chen Y-CA, Godneeva B, Rogers AK, Luo Y, Fejes Tóth K, Aravin AA (2020) Su(var)2–10 and the SUMO pathway link piRNA-guided target recognition to chromatin silencing. Mol Cell. https://doi.org/10.1016/j.molcel.2019.11.012
Wolf G, Greenberg D, Macfarlan TS (2015) Spotting the enemy within: targeted silencing of foreign DNA in mammalian genomes by the Krüppel-associated box zinc finger protein family. Mob DNA 6:17. https://doi.org/10.1186/s13100-015-0050-8
Osumi K, Sato K, Murano K, Siomi H, Siomi MC (2019) Essential roles of Windei and nuclear monoubiquitination of Eggless/SETDB1 in transposon silencing. EMBO Rep 20(12):e48296. https://doi.org/10.15252/embr.201948296
Stein P, Rozhkov NV, Li F, Cárdenas FL, Davydenko O, Davydenk O, Vandivier LE, Gregory BD, Hannon GJ, Schultz RM (2015) Essential role for endogenous siRNAs during meiosis in mouse oocytes. PLoS Genet 11(2):e1005013. https://doi.org/10.1371/journal.pgen.1005013
Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ (2008) Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453(7194):534–538. https://doi.org/10.1038/nature06904
Taborska E, Pasulka J, Malik R, Horvat F, Jenickova I, Jelić Matošević Z, Svoboda P (2019) Restricted and non-essential redundancy of RNAi and piRNA pathways in mouse oocytes. PLoS Genet 15(12):e1008261. https://doi.org/10.1371/journal.pgen.1008261
Yamashiro H, Negishi M, Kinoshita T, Ishizu H, Ohtani H, Siomi MC (2020) Armitage determines Piwi-piRISC processing from precursor formation and quality control to inter-organelle translocation. EMBO Rep 21(2):e48769. https://doi.org/10.15252/embr.201948769
Ishizu H, Kinoshita T, Hirakata S, Komatsuzaki C, Siomi MC (2019) Distinct and collaborative functions of Yb and armitage in transposon-targeting piRNA biogenesis. Cell Rep. https://doi.org/10.1016/j.celrep.2019.04.029
Kordyukova M, Sokolova O, Morgunova V, Ryazansky S, Akulenko N, Glukhov S, Kalmykova A (2020) Nuclear Ccr4-Not mediates the degradation of telomeric and transposon transcripts at chromatin in the Drosophila germline. Nucleic Acids Res 48(1):141–156. https://doi.org/10.1093/nar/gkz1072
Dai P, Wang X, Gou L-T, Li Z-T, Wen Z, Chen Z-G, Hua M-M, Zhong A, Wang L, Su H, Wan H, Qian K, Liao L, Li J, Tian B, Li D, Fu X-D, Shi H-J, Zhou Y, Liu M-F (2019) A translation-activating function of MIWI/piRNA during mouse spermiogenesis. Cell. https://doi.org/10.1016/j.cell.2019.11.022
Ophinni Y, Palatini U, Hayashi Y, Parrish NF (2019) piRNA-guided CRISPR-like immunity in eukaryotes. Trends Immunol. https://doi.org/10.1016/j.it.2019.09.003
Yu T, Koppetsch BS, Pagliarani S, Johnston S, Silverstein NJ, Luban J, Chappell K, Weng Z, Theurkauf WE (2019) The piRNA response to retroviral invasion of the koala genome. Cell. https://doi.org/10.1016/j.cell.2019.09.002
Mao Q, Fan L, Wang X, Lin X, Cao Y, Zheng C, Zhang Y, Zhang H, Garcia-Milian R, Kang L, Shi J, Yu T, Wang K, Zuo L, Li C-SR, Guo X, Luo X (2019) Transcriptome-wide piRNA profiling in human brains for aging genetic factors. Jacobs J Genet 4(1):14
Mai D, Ding P, Tan L, Zhang J, Pan Z, Bai R, Li C, Li M, Zhou Y, Tan W, Zhou Z, Li Y, Zhou A, Ye Y, Pan L, Zheng Y, Su J, Zuo Z, Liu Z, Zhao Q, Li X, Huang X, Li W, Wu S, Jia W, Zou S, Wu C, Xu R-H, Zheng J, Lin D (2018) PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics 8(19):5213–5230. https://doi.org/10.7150/thno.28001
Liu Y, Dou M, Song X, Dong Y, Liu S, Liu H, Tao J, Li W, Yin X, Xu W (2019) The emerging role of the piRNA/piwi complex in cancer. Mol Cancer 18(1):123. https://doi.org/10.1186/s12943-019-1052-9
Ai L, Mu S, Sun C, Fan F, Yan H, Qin Y, Cui G, Wang Y, Guo T, Mei H, Wang H, Hu Y (2019) Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing piRNA-823 expression and DNMT3B activation. Mol Cancer 18(1):88. https://doi.org/10.1186/s12943-019-1011-5
Liu Y, Li G, Lu H, Li W, Li X, Liu H, Li X, Li T, Yu B (2014) Expression profiling and ontology analysis of long noncoding RNAs in post-ischemic heart and their implied roles in ischemia/reperfusion injury. Gene 543(1):15–21. https://doi.org/10.1016/j.gene.2014.04.016
Wang J-X, Zhang X-J, Li Q, Wang K, Wang Y, Jiao J-Q, Feng C, Teng S, Zhou L-Y, Gong Y, Zhou Z-X, Liu J, Wang J-L, Li P-f (2015) MicroRNA-103/107 regulate programmed necrosis and myocardial ischemia/reperfusion injury through targeting FADD. Circ Res 117(4):352–363. https://doi.org/10.1161/CIRCRESAHA.117.305781
Zhao LY, Zhang J, Guo B, Yang J, Han J, Zhao XG, Wang XF, Liu LY, Li ZF, Song TS, Huang C (2013) MECP2 promotes cell proliferation by activating ERK1/2 and inhibiting p38 activity in human hepatocellular carcinoma HEPG2 cells. Cell Mol Biol (Noisy-le-grand) 59:OL1876–OL1881
Law PTY, Qin H, Ching AKK, Lai KP, Co NN, He M, Lung RWM, Chan AWH, Chan T-F, Wong N (2013) Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol 58(6):1165–1173. https://doi.org/10.1016/j.jhep.2013.01.032
Duan L, Hu J, Xiong X, Liu Y, Wang J (2018) The role of DNA methylation in coronary artery disease. Gene 646:91–97. https://doi.org/10.1016/j.gene.2017.12.033
Kızıltunç E, Kösem A, Özkan C, Ilgın BU, Kundi H, Çetin M, Ornek E (2019) Serum sirtuin 1, 3 and 6 levels in acute myocardial infarction patients. Arq Bras Cardiol 113(1):33–39. https://doi.org/10.5935/abc.20190114
Ottaviani L, da Costa Martins PA (2017) Non-coding RNAs in cardiac hypertrophy. J Physiol (Lond) 595(12):4037–4050. https://doi.org/10.1113/JP273129
Peng L, Song L, Liu C, Lv X, Li X, Jie J, Zhao D, Li D (2016) piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling. Tumour Biol 37(2):2749–2756. https://doi.org/10.1007/s13277-015-4056-0
Qi H, Ren J, Ba L, Song C, Zhang Q, Cao Y, Shi P, Fu B, Liu Y, Sun H (2020) MSTN attenuates cardiac hypertrophy through inhibition of excessive cardiac autophagy by blocking AMPK /mTOR and miR-128/PPARγ/NF-κB. Mol Ther Nucleic Acids 19:507–522. https://doi.org/10.1016/j.omtn.2019.12.003
Crippa S, Nemir M, Ounzain S, Ibberson M, Berthonneche C, Sarre A, Boisset G, Maison D, Harshman K, Xenarios I, Diviani D, Schorderet D, Pedrazzini T (2016) Comparative transcriptome profiling of the injured zebrafish and mouse hearts identifies miRNA-dependent repair pathways. Cardiovasc Res 110(1):73–84. https://doi.org/10.1093/cvr/cvw031
Chong ZZ, Shang YC, Maiese K (2011) Cardiovascular disease and mTOR signaling. Trends Cardiovasc Med 21(5):151–155. https://doi.org/10.1016/j.tcm.2012.04.005
Pipicz M, Demján V, Sárközy M, Csont T (2018) Effects of cardiovascular risk factors on cardiac STAT3. Int J Mol Sci. https://doi.org/10.3390/ijms19113572
Aplin AC, Nicosia RF (2019) The plaque-aortic ring assay: a new method to study human atherosclerosis-induced angiogenesis. Angiogenesis 22(3):421–431. https://doi.org/10.1007/s10456-019-09667-z
Wu Q, Xu W-D, Huang A-F (2020) Role of angiopoietin-2 in inflammatory autoimmune diseases: a comprehensive review. Int Immunopharmacol 80:106223. https://doi.org/10.1016/j.intimp.2020.106223
Xian D, Song J, Yang L, Xiong X, Lai R, Zhong J (2019) Emerging roles of redox-mediated angiogenesis and oxidative stress in dermatoses. Oxid Med Cell Longev 2019:2304018. https://doi.org/10.1155/2019/2304018
Cheng J, Guo J-M, Xiao B-X, Miao Y, Jiang Z, Zhou H, Li Q-N (2011) piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta 412(17–18):1621–1625. https://doi.org/10.1016/j.cca.2011.05.015
Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J (2012) piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett 315(1):12–17. https://doi.org/10.1016/j.canlet.2011.10.004
Yan H, Wu QL, Sun CY, Ai LS, Deng J, Zhang L, Chen L, Chu ZB, Tang B, Wang K, Wu XF, Xu J, Hu Y (2015) piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia 29(1):196–206. https://doi.org/10.1038/leu.2014.135
Li B, Hong J, Hong M, Wang Y, Yu T, Zang S, Wu Q (2019) piRNA-823 delivered by multiple myeloma-derived extracellular vesicles promoted tumorigenesis through re-educating endothelial cells in the tumor environment. Oncogene 38(26):5227–5238. https://doi.org/10.1038/s41388-019-0788-4
Zhang X, Huang F, Li W, Dang J-L, Yuan J, Wang J, Zeng D-L, Sun C-X, Liu Y-Y, Ao Q, Tan H, Su W, Qian X, Olsen N, Zheng SG (2018) Human gingiva-derived mesenchymal stem cells modulate monocytes/macrophages and alleviate atherosclerosis. Front Immunol 9:878. https://doi.org/10.3389/fimmu.2018.00878
Zhang X, Huang F, Chen Y, Qian X, Zheng SG (2016) Progress and prospect of mesenchymal stem cell-based therapy in atherosclerosis. Am J Transl Res 8(10):4017–4024
Li J, Xue H, Li T, Chu X, Xin D, Xiong Y, Qiu W, Gao X, Qian M, Xu J, Wang Z, Li G (2019) Exosomes derived from mesenchymal stem cells attenuate the progression of atherosclerosis in ApoE mice via miR-let7 mediated infiltration and polarization of M2 macrophage. Biochem Biophys Res Commun 510(4):565–572. https://doi.org/10.1016/j.bbrc.2019.02.005
Moghaddam AS, Afshari JT, Esmaeili S-A, Saburi E, Joneidi Z, Momtazi-Borojeni AA (2019) Cardioprotective microRNAs: lessons from stem cell-derived exosomal microRNAs to treat cardiovascular disease. Atherosclerosis 285:1–9. https://doi.org/10.1016/j.atherosclerosis.2019.03.016
Xing X, Li Z, Yang X, Li M, Liu C, Pang Y, Zhang L, Li X, Liu G, Xiao Y (2020) Adipose-derived mesenchymal stem cells-derived exosome-mediated microRNA-342-5p protects endothelial cells against atherosclerosis. Aging (Albany NY) 12(4):3880–3898. https://doi.org/10.18632/aging.102857
Liu S, Yang Y, Jiang S, Tang N, Tian J, Ponnusamy M, Tariq MA, Lian Z, Xin H, Yu T (2018) Understanding the role of non-coding RNA (ncRNA) in stent restenosis. Atherosclerosis 272:153–161. https://doi.org/10.1016/j.atherosclerosis.2018.03.036
Li M, Ding W, Tariq MA, Chang W, Zhang X, Xu W, Hou L, Wang Y, Wang J (2018) A circular transcript of gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics 8(21):5855–5869. https://doi.org/10.7150/thno.27285
Yang Y, Yu T, Jiang S, Zhang Y, Li M, Tang N, Ponnusamy M, Wang J-X, Li P-F (2017) miRNAs as potential therapeutic targets and diagnostic biomarkers for cardiovascular disease with a particular focus on WO2010091204. Expert Opin Ther Pat 27(9):1021–1029. https://doi.org/10.1080/13543776.2017.1344217
Bei Y, Yang T, Wang L, Holvoet P, Das S, Sluijter JPG, Monteiro MC, Liu Y, Zhou Q, Xiao J (2018) Circular RNAs as potential theranostics in the cardiovascular system. Mol Ther Nucleic Acids 13:407–418. https://doi.org/10.1016/j.omtn.2018.09.022
Mompeón A, Ortega-Paz L, Vidal-Gómez X, Costa TJ, Pérez-Cremades D, Garcia-Blas S, Brugaletta S, Sanchis J, Sabate M, Novella S, Dantas AP, Hermenegildo C (2020) Disparate miRNA expression in serum and plasma of patients with acute myocardial infarction: a systematic and paired comparative analysis. Sci Rep 10(1):5373. https://doi.org/10.1038/s41598-020-61507-z
Wen P, Song D, Ye H, Wu X, Jiang L, Tang B, Zhou Y, Fang L, Cao H, He W, Yang Y, Dai C, Yang J (2014) Circulating MiR-133a as a biomarker predicts cardiac hypertrophy in chronic hemodialysis patients. PLoS ONE 9(10):e103079. https://doi.org/10.1371/journal.pone.0103079
Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442(7099):203–207
Acknowledgements
This work was supported by The National Natural Science Foundation of China (Grant No. 81870331, 31701208), The Natural Science Foundation of Shandong Province (Grant No. ZR2017MC067), and The Qingdao municipal science and technology bureau project (Grant No. 18-2-2-65-jch).
Author information
Authors and Affiliations
Contributions
ML, YY, and ZW collected materials and wrote the manuscript. TY and JW provided the idea. KW and TZ are responsible for the schematic diagram within this article. TY, LHHA, and XF helped with the final revision of the article. All authors reviewed the manuscript and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that they have no competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, M., Yang, Y., Wang, Z. et al. Piwi-interacting RNAs (piRNAs) as potential biomarkers and therapeutic targets for cardiovascular diseases. Angiogenesis 24, 19–34 (2021). https://doi.org/10.1007/s10456-020-09750-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10456-020-09750-w