Abstract
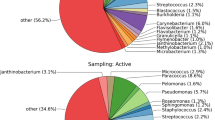
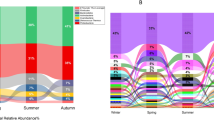
As the main biological components of the air ecosystem, airborne microorganisms play an important role in ecosystem function, and most existing studies focus on their health and environmental effects, while ignoring ecological effects. In this study, air microbial samples from two sampling points in the coastal city of Qingdao were collected by a large volume air sampler. The Illumina MiSeq and PacBio high-throughput sequencing methods were used to analyze the microbial community structure and diversity of the two sampling points. The results showed that there were significant differences in the number and relative abundances of microorganisms identified by the two platforms, and with the refinement of the sample classification, the differences in the community became increasingly significant. Proteobacteria and Firmicutes were the dominant bacterial phyla among the airborne microorganisms. MiSeq sequencing showed that the relative abundances of unidentified bacteria were as high as 8.03–16.42% in MiSeq sequencing, while they were almost undetected in PacBio sequencing. Based on the different sequencing platforms, Bacilli, Gammaproteobacteria and Alphaproteobacteria could be seen as significant gaps in the two sampling points. Betaproteobacteria showed differences only in the coastal area, while the other 14 classes were differentially abundant in the urban area. Pseudomonas was the main genus in the aerosol, followed by Lactococcus. The relative abundances of bacterial taxa displayed statistically significant differences in the different functional areas. There were more diverse microbes in the urban area than in the coastal area; moreover, the relative abundances of dominant genera showed significant differences, such as Variovorax and Deinococcus.






Similar content being viewed by others
References
Bertolini, V., Gandolfi, I., Ambrosini, R., Bestetti, G., Innocente, E., Rampazzo, G., et al. (2013). Temporal variability and effect of environmental variables on airborne bacterial communities in an urban area of Northern Italy. Applied Microbiology and Biotechnology, 97, 6561–6570.
Bowers, R. M., Mcletchie, S., Knight, R., & Fierer, N. (2011). Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. The ISME Journal, 5, 601–612.
Bragoszewska, E., & Biedroń, I. (2018). Indoor air quality and potential health risk impacts of exposure to antibiotic resistant bacteria in an office rooms in Southern Poland. International Journal of Environmental Research and Public Health, 15, 139–149.
Bruce, S. A., Saville, B. J., & Emery, R. J. N. (2011). Ustilago maydis produces cytokinins and abscisic acid for potential regulation of tumor formation in maize. Journal of Plant Growth Regulation, 30, 51–63.
Daneshvar, M. I., Hollis, D. G., Weyant, R. S., Steigerwalt, A. G., Whitney, A. M., Douglas, M. P., et al. (2003). Paracoccus yeeii sp. nov. (formerly cdc group eo-2), a novel bacterial species associated with human infection. Journal of Clinical Microbiology, 41, 1289–1294.
Derakhshani, H., Tun, H. M., & Khafipour, E. (2016). An extended single-index multiplexed 16S rRNA sequencing for microbial community analysis on MiSeq illumina platforms. Journal of Basic Microbiology, 56, 321–326.
Doughari, H. J., Ndakidemi, P. A., Human, I. S., & Benade, S. (2011). The ecology, biology and pathogenesis of Acinetobacter spp.: An overview. Microbes and Environments, 26, 101–112.
Du, P. R., Du, R., Ren, W. S., Lu, Z. D., Zhang, Y., & Fu, P. Q. (2018). Variations of bacteria and fungi in PM2.5, in Beijing, China. Atmospheric Environment, 172, 55–64.
Fahlgren, C., Bratbak, G., Sandaa, R. A., Thyrhaug, R., & Zweifel, U. L. (2011). Diversity of airborne bacteria in samples collected using different devices for aerosol collection. Aerobiologia, 27, 107–120.
Fahlgren, C., Hagstrom, A., Nilsson, D., & Zweifel, U. L. (2010). Annual variations in the diversity, viability, and origin of airborne bacteria. Applied and Environmental Microbiology, 76, 3015–3025.
Frellsen, J., Menzel, P., & Krogh, A. (2014). Algorithms for mapping high-throughput DNA sequences. Comprehensive Biomedical Physics, 57, 41–50.
Gao, Q., Yue, G. D., Li, W. Q., Wang, J. Y., Xu, J. H., & Yin, Y. (2012). Recent Progress using high-throughput sequencing technologies in plant molecular breeding. Journal of Integrative Plant Biology, 54, 215–227.
Gou, H., Lu, J., Li, S., Tong, Y., Xie, C., & Zheng, X. (2016). Assessment of microbial communities in PM1 and PM10 of Uru’mqi during winter. Environmental Pollution, 214, 202–210.
Griffin, D. W., Kubilay, N., Koçak, M., Gray, M. A., Borden, T. C., & Shinn, E. A. (2007). Airborne desert dust and aeromicrobiology over the Turkish mediterranean coastline. Atmospheric Environment, 41, 4050–4062.
Innocente, E., Squizzato, S., Visin, F., Facca, C., Rampazzo, G., Bertolini, V., et al. (2017). Influence of seasonality, air mass origin and particulate matter chemical composition on airborne bacterial community structure in the Po Valley, Italy. Science of The Total Environment, 593–594, 677–687.
Jeon, E. M., Kim, H. J., Jung, K., Kim, J. H., Kim, M. Y., Kim, Y. P., et al. (2011). Impact of Asian dust events on airborne bacterial community assessed by molecular analyses. Atmospheric Environment, 45, 4313–4321.
Kaushik, R., & Balasubramanian, R. (2012). Assessment of bacterial pathogens in fresh rainwater and airborne particulate matter using real-time PCR. Atmospheric Environment, 46, 131–139.
Khiste, N., & Ilie, L. (2017). Hisea: hierarchical seed aligner for Pacbio data. BMC Bioinformatics, 18, 564.
Kim, S. B., Nedashkovskaya, O. I., Mikhailov, V. V., Han, S. K., & Bae, K. S. (2004). Kocuria marina sp. nov. a novel actinobacterium isolated from marine sediment. International Journal of Systematic and Evolutionary Microbiology, 54, 1617–1620.
Koichi, Y., Yoshitomo, M., Katsunori, Y., Norihito, K., Yosuke, H., Naoki, U., et al. (2014). Azithromycin inhibits MUC5AC induction via multidrug-resistant Acinetobacter baumannii in human airway epithelial cells. Pulmonary Pharmacology & Therapeutics, 28, 165–170.
Kucisec-Tepes N, Bejuk D, & Kosuta, D. (2006). [characteristics of war wound infection]. Acta medica Croatica: časopis Hravatske akademije medicinskih znanosti, 60, 353–363.
Lee, S. H., Lee, H. J., Kim, S. J., Lee, H. M., Kang, H., & Kim, Y. P. (2010). Identification of airborne bacterial and fungal community structures in an urban area by t-RFLP analysis and quantitative real-time PCR. Science of the Total Environment, 408, 1349–1357.
Li, Y. P., Fu, H. L., Wang, W., Liu, J., Meng, Q. L., & Wang, W. K. (2015). Characteristics of bacterial and fungal aerosols during the autumn haze days in Xi’an, China. Atmospheric Environment, 122, 439–447.
Lu, H., Fujimura, R., Sato, Y., Nanba, K., Kamijo, T., & Ohta, H. (2008). Characterization of herbaspirillum- and limnobacter-related strains isolated from young volcanic deposits in miyake-jima island, Japan. Microbes and Environments, 23, 66–72.
Lu, R., Li, Y. P., Li, W. X., Xie, Z. S., Fan, C. L., Liu, P. X., et al. (2018). Bacterial community structure in atmospheric particulate matters of different sizes during the haze days in Xi’an, China. Science of The Total Environment, 637–638, 244–252.
Magoc, T., & Salzberg, S. L. (2011). Flash: fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 27, 2957–2963.
Meyer, C., Bernard, N., Moskura, M., Toussaint, M. L., Denayer, F., & Gilbert, D. (2010). Effects of urban particulate deposition on microbial communities living in bryophytes: An experimental study. Ecotoxicology and Environmental Safety, 73, 1776–1784.
Munday, C., De-Deckker, P., Tapper, N., O’Loingsigh, T., & Allison, G. (2016). Characterizing bacterial assemblages in sediments and aerosols at a dry lake bed in australia using high-throughput sequencing. Aerobiologia, 32, 581–593.
Pylro, V. S., Roesch, L. F. W., Morais, D. K., Clark, I. M., Hirsch, P. R., & Tótola, M. R. (2014). Data analysis for 16s microbial profiling from different benchtop sequencing platforms. Journal of Microbiological Methods, 107, 30–37.
Qi, J. H., Li, M. Z., Zhe, N. Y., & Wu, L. J. (2018). Characterization of bioaerosol bacterial communities during hazy and foggy weather in Qingdao, China. Journal of Ocean University of China, 17, 71–81.
Rindi, F. (2007). Diversity, distribution and ecology of green algae and cyanobacteria in urban habitats. Cellular Origin Life in Extreme Habitats & Astrobiology, 11, 619–638.
Robertson, C. E., Baumgartner, L. K., Harris, J. K., Peterson, K. L., Stevens, M. J., Frank, D. N., et al. (2013). Culture-independent analysis of aerosol microbiology in a metropolitan subway system. Applied & Environmental Microbiology, 79, 3485–3493.
Robine, E., Dérangère, D., & Robin, D. (2000). Survival of a pseudomonas fluorescens and Enterococcus faecalis aerosol on inert surfaces. International Journal of Food Microbiology, 55, 229–234.
Smith, D. J., Timonen, H. J., Jaffe, D. A., Griffin, D. W., Birmele, M. N., Perry, K. D., et al. (2013). Intercontinental dispersal of bacteria and archaea by transpacific winds. Applied and Environmental Microbiology, 79, 1134–1139.
Spring, S., Kämpfer, P., & Schleifer, K. H. (2001). Limnobacter thiooxidans gen nov. sp. Nov. a novel thiosulfate-oxidizing bacterium isolated from freshwater lake sediment. International Journal of Systematic and Evolutionary Microbiology, 51, 1463–1470.
Sun, Y., Xu, S., Zheng, D., Li, J., Tian, H., & Wang, Y. (2018). Effects of haze pollution on microbial community changes and correlation with chemical components in atmospheric particulate matter. Science of the Total Environment, 637–638, 507.
Szponar, B., & Larsson, L. (2001). Use of mass spectrometry for characterising microbial communities in bioaerosols. Annals of Agricultural and Environmental Medicine: AAEM, 8, 111–117.
Tanaka, D., Terada, Y., Nakashima, T., Sakatoku, A., & Nakamura, S. (2015). Seasonal variations in airborne bacterial community structures at a suburban site of central Japan over a 1-year time period using PCR-DGGE method. Aerobiologia, 31, 143–157.
Tang, K., Lin, Y. F., Han, Y., & Jiao, N. Z. (2017). Characterization of potential polysaccharide utilization systems in the marine Bacteroidetes Gramella Flava JLT2011 using a multi-omics approach. Frontiers in Microbiology, 8, 220.
Unno, T. (2015). Bioinformatic suggestions on miseq-based microbial community analysis. Journal of Microbiology and Biotechnology, 25, 765.
Weon, H. Y., Son, J. A., Yoo, S. H., Hong, S. B., Jeon, Y. A., Kwon, S. W., et al. (2009). Rubellimicrobium aerolatum sp. nov. isolated from an air sample in Korea. International Journal of Systematic and Evolutionary Microbiology, 59, 406–410.
Whon, T. W., Chung, W. H., Lim, M. Y., Song, E. J., Kim, P. S., Hyun, D. W., et al. (2018). The effects of sequencing platforms on phylogenetic resolution in 16 S rRNA gene profiling of human feces. Scientific Data, 5, 180068.
Yeo, H. G., & Kim, J. H. (2002). SPM and fungal spores in the ambient air of west korea during the asian dust (Yellow sand) period. Atmospheric Environment, 36, 5437–5442.
Yu, M., Sun, Q., Song, Z. W., & Wu, L. (2010). Investigation of microbial aerosol in a full-scale surface flow constructed wetland. Environmental Pollution And Prevention, 32, 8–12.
Zahradka, K., Slade, D., Bailone, A., Sommer, S., Averbeck, D., Petranovic, M., et al. (2006). Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature, 443, 569–573.
Zevenboom, W. (1990). Picocyanobacteria in the Banda Sea during two different monsoons. Netherlands Journal of Sea Research, 25, 513–521.
Zhang, Y. Y., Du, M. M., Chang, Y., Chen, L. A., & Zhang, Q. (2017). Incidence, clinical characteristics, and outcomes of Nosocomial enterococcus spp. bloodstream infections in a tertiary-care hospital in Beijing, China: A four-year retrospective study. Antimicrobial Resistance & Infection Control, 6, 73.
Zweifel, U. L., Hagström, Å., Holmfeldt, K., Thyrhaug, R., Geels, C., Frohn, L. M., et al. (2012). High bacterial 16s rRNA gene diversity above the atmospheric boundary layer. Aerobiologia, 28, 481–498.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (No. 31570541, 31170509). We sincerely thank the staff at WWTPs of Jiaonan, Shandong Province, China for their help with sampling. The authors also thank other members of our laboratory who provided their valuable and constructive suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lang, Xl., Xu, Al., Chen, X. et al. Aerosol microbial community structure analysis based on two different sequencing platforms. Aerobiologia 36, 617–630 (2020). https://doi.org/10.1007/s10453-020-09655-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10453-020-09655-7