Abstract
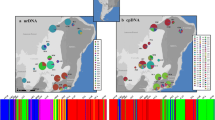
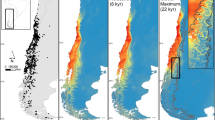
Several evolutionary processes seem to have influenced the Atlantic Forest (AF) biogeographic history, as suggested by phylogeographic studies that have shown a multitude of patterns. Here, we use approximate Bayesian computation to test alternative historical hypotheses to investigate the phylogeographic pattern, historical demography, and palaeodistribution of the Grey-hooded Flycatcher Mionectes rufiventris, an endemic AF bird, distributed mainly in southern areas of the biome. Our goal was to integrate molecular and ecological data to test diversification hypotheses available for the AF. Our investigation revealed two mitochondrial phylogroups, geographically structured around the Doce River. Coalescence analyses revealed that these groups shared a common ancestor in the Late Pleistocene, between 200,000 and 300,000 years ago, and that divergence was probably associated with climatic fluctuations during this period. Demographic analyses suggested recent demographic expansion in both groups. Ecological niche modelling suggested larger ranges during the Last Glacial Maximum (LGM) than in the present, not in agreement with the genetic pattern recovered. We simulated alternative historical models to test these competing scenarios. Our findings support the existence of small populations during the LGM which expanded afterwards from putative refuges. Thus, these results suggest that the Pleistocene climate shaped patterns of diversification and demographic history of this species in accordance with the classical forest refuge hypothesis.
Zusammenfassung
Klimaänderungen im späten Pleistozän prägten die Populationsdivergenz eines Singvogels der Mata Atlantica: ein modellbasierter Test phylogeografischer Hypothesen
Mehrere evolutionäre Prozesse scheinen die biogeografische Geschichte der Mata Atlantica (engl.: Atlantic Forest, AF) beeinflusst zu haben, wie phylogeografische Studien nahelegen, welche eine Unzahl von Mustern aufgedeckt haben. Mittels Bayes’scher Näherungsrechnung (engl.: Approximate Bayesian Computation, ABC) prüften wir alternative historische Hypothesen, um die phylogeografischen Muster, die historische Demografie sowie die paläologische Verbreitung des Graukopf-Pipratyrann Mionectes rufiventris zu untersuchen, einer für den AF endemischen Vogelart, welche überwiegend in den südlichen Bereichen dieses Biomes verbreitet ist. Unser Ziel war es, unter Einbeziehung molekularer und ökologischer Daten die für den AF verfügbaren Diversifikationshypothesen zu prüfen. Unsere Multilokus-Untersuchung ließ zwei phylogenetische Gruppen erkennen, die geografisch um den Rio Doce angeordnet waren. Koalenszenzanalysen zeigten, dass diese Gruppen einen gemeinsamen Vorfahren im späten Pleistozän (vor 200.000-300.000 Jahren) besaßen und dass die Divergenz vermutlich mit Klimafluktuationen während dieser Periode zusammenhing. Demografische Analysen deuteten auf eine vor kurzem erfolgte demografische Expansion in beiden Gruppen hin. Die ökologische Nischenmodellierung ließ vermuten, dass die Verbreitungsgebiete während des Letzteiszeitlichen Maximums (engl.: Last Glacial Maximum, LGM) größer waren als heute, was nicht mit dem beobachteten genetischen Muster in Einklang steht. Zur Prüfung dieser konkurrierenden Szenarien führten wir Simulationen alternativer historischer Modelle durch. Unsere Befunde sprechen für die Existenz kleiner Populationen während des LGM, die sich anschließend ausgehend von angenommenen Refugien ausbreiteten. Diese Ergebnisse deuten somit darauf hin, dass das Klima im Pleistozän die Diversitätsmuster und die demografische Geschichte dieser Vogelart gemäß der klassischen Waldrefugien-Hypothese gestaltet hat.



Similar content being viewed by others
References
Amaro RC, Rodrigues MT, Yonenaga-Yassuda Y, Carnaval AC (2012) Demographic processes in the montane Atlantic rainforest: molecular and cytogenetic evidence from the endemic frog Proceratophrys boiei. Mol Phylogenet Evol 62:880–888
Araújo MB, New M (2007) Ensemble forecasting of species distributions. Trends Ecol Evol 22:42–47
Batalha-Filho H, Miyaki CY (2016) Late Pleistocene divergence and postglacial expansion in the Brazilian Atlantic Forest: multilocus phylogeography of Rhopias gularis (Aves: Passeriformes). J Zool Syst Evol Res 54:137–147
Batalha-Filho H, Waldschmidt AM, Campos LA, Tavares MG, Fernandes-Salomão TM (2010) Phylogeography and historical demography of the Neotropical stingless bee Melipona quadrifasciata (Hymenoptera, Apidae): incongruence between morphology and mitochondrial DNA. Apidologie 41:534–547
Batalha-Filho H, Cabanne GS, Miyaki CY (2012) Phylogeography of an Atlantic forest passerine reveals demographic stability through the last glacial maximum. Mol Phylogenet Evol 65:892–902
Behling H (2002) South and southeast Brazilian grasslands during Late Quaternary times: a synthesis. Palaeogeogr Palaeoclimatol Palaeoecol 177:19–27
Behling H, Pillar VD, Bauermann SG (2005) Late Quaternary grassland (Campos), gallery forest, fire and climate dynamics, studied by pollen, charcoal and multivariate analysis of the São Francisco de Assis core in western Rio Grande do Sul (southern Brazil). Rev Palaeobot Palynol 133:235–248
Britto PH, Edwards SV (2009) Multilocus phylogeography and phylogenetics using sequence-based markers. Genetica 135:439–455
Bruen TC, Philippe H, Bryant D (2006) A simple and robust statistical test for detecting the present of recombination. Genetics 172:2665–2681
Brunes TO, Thomé MTC, Alexandrino J, Haddad CF, Sequeira F (2015) Ancient divergence and recent population expansion in a leaf frog endemic to the southern Brazilian Atlantic forest. Org Divers Evol 15:695–710
Cabanne GS, Santos FR, Miyaki CY (2007) Phylogeography of Xiphorhynchus fuscus (Passeriformes, Dendrocolaptidae): vicariance and recent demographic expansion in southern Atlantic forest. Biol J Linn Soc Lond 91:73–84
Cabanne GS, d’Horta FM, Sari EH, Santos FR, Miyaki CY (2008) Nuclear and mitochondrial phylogeography of the Atlantic forest endemic Xiphorhynchus fuscus (Aves: Dendrocolaptidae): biogeography and systematics implications. Mol Phylogenet Evol 49:760–773
Cabanne GS, Sari EH, Meyer D, Santos FR, Miyaki CY (2013) Matrilineal evidence for demographic expansion, low diversity and lack of phylogeographic structure in the Atlantic forest endemic Greenish Schiffornis Schiffornis virescens (Aves: Tityridae). J Ornithol 154:371–384
Cabanne GS, Calderón L, Arias NT, Flores P, Pessoa R, d’Horta FM et al (2016) Effects of Pleistocene climate changes on species ranges and evolutionary processes in the Neotropical Atlantic Forest. Biol J Linn Soc Lond 119:856–872
Carnaval AC, Moritz C (2008) Historical climate modelling predicts patterns of current biodiversity in the Brazilian Atlantic forest. J Biogeogr 35:1187–1201
Carnaval AC, Hickerson MJ, Haddad CF, Rodrigues MT, Moritz C (2009) Stability predicts genetic diversity in the Brazilian Atlantic forest hotspot. Science 323:785–789
Carnaval AC, Waltari E, Rodrigues MT, Rosauer D, VanDerWal J, Damasceno R et al (2014) Prediction of phylogeographic endemism in an environmentally complex biome. Proc R Soc Lond B Biol Sci 281:20141461
Clapperton C (1993) Quaternary geology and geomorphology of South America. Elsevier, New York, pp 143–162
Collevatti RG, Terribile LC, Diniz-Filho JA, Lima-Ribeiro MS (2015) Multi-model inference in comparative phylogeography: an integrative approach based on multiple lines of evidence. Front Genet 6:31
Csilléry K, François O, Blum MGB (2012) abc: an R package for approximate Bayesian computation (ABC). Methods Ecol Evol 3:475–479
d’Horta FM, Cabanne GS, Meyer D, Miyaki CY (2011) The genetic effects of Late Quaternary climatic changes over a tropical latitudinal gradient: diversification of an Atlantic Forest passerine. Mol Ecol 20:1923–1935
Darriba D, Taboada GL, Doallo R, Posada D (2012) Jmodeltest 2: more models, new heuristics and parallel computing. Nat Methods 9:772
Del Hoyo J, Elliott A, Sargatal J, Christie DA (2004) Handbook of the birds of the world—vol 9: cotingas to pipits and wagtails, 1st edn. Ed. Lynx Edicions, Barcelona
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214
Edwards SV, Beerli P (2000) Perspective: gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies. Evolution 54:1839–1854
Ellegren H (2007) Molecular evolutionary genomics of birds. Cytogenet Genome Res 117:120–130
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Fitzpatrick SW, Brasileiro CA, Haddad CFB, Zamudio KR (2009) Geographical variation in genetic structure of an Atlantic Coastal Forest frog reveals regional differences in habitat stability. Mol Ecol 18:2877–2896
Friesen VL, Congdon BC, Walsh HE, Birt TP (1997) Intron variation in marbled murrelets detected using analyses of single-stranded conformational polymorphisms. Mol Ecol 6:1047–1058
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Guillot G, Estoup A, Mortier F, Cosson JF (2005) A spatial statistical model for landscape genetics. Genetics 170:1261–1280
Haffer J (1969) Speciation in Amazonian forest birds. Science 165:131–137
Helen J, Drummond AJ (2008) Bayesian inference of population size history from multiple loci. BMC Evol Biol 8:289
Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
Huang W, Takebayashi N, Qi Y, Hickerson MJ (2011) MTML-msBayes: approximate Bayesian comparative phylogeographic inference from multiple taxa and multiple loci with rate heterogeneity. BMC Bioinform 12:1
Huson DH, Bryant D (2006) Applications of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Jensen JL, Bohonak AJ, Kelley S (2005) Isolation by distance, web service. BMC Genet 6:13 v.3.23. http://ibdws.sdsu.edu/
Knowles LL (2009) Statistical phylogeography. Annu Rev Ecol Evol Syst 40:593–612
Kuhner MK (2006) LAMARC 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics 22:768–770
Ledru MP, Salgado-Labouriau ML, Lorscheitter ML (1998) Vegetational dynamics in southern and central Brazil during the last 10,000 yr B.P. Rev Palaeobot Palynol 99:131–142
Leigh JW, Bryant D (2015) POPART: full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116
Leite YL, Costa LP, Loss AC, Rocha RG, Batalha-Filho H, Bastos AC et al (2016) Neotropical forest expansion during the last glacial period challenges refuge hypothesis. Proc Natl Acad Sci USA 113:1008–1013
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lougheed SC, Freeland JR, Handford P, Boag PT (2000) A molecular phylogeny of warbling-finches (Poospiza): paraphyly in a Neotropical emberizid genus. Mol Phylogenet Evol 17:367–378
Maldonado-Coelho M (2012) Climatic oscillations shape the phylogeographical structure of Atlantic Forest fire-eye antbirds (Aves: Thamnophilidae). Biol J Linn Soc 105:900–924
Martins FM, Gifalli-Iughetti C, Koiffman CP, Harris EE (2011) Coalescent analysis of mtDNA indicates Pleistocene divergence among three species of howler monkey (Alouatta spp.) and population subdivision within the Atlantic Coastal Forest species A. guariba. Primates 52:77–87
McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654
Miller MJ, Bermingham E, Klicka J, Escalante P, Amaral FSR, Weir JT, Winker K (2008) Out of Amazonia again and again: episodic crossing of the Andes promotes diversification in a lowland forest flycatcher. Proc R Soc B 275:1133–1142
Moritz C, Patton JL, Schneider CJ, Smith TB (2000) Diversification of rainforest faunas: an integrated molecular approach. Annu Rev Ecol Syst 31:533–563
Muhs DR, Simmons KR, Steinke B (2002) Timing and warmth of the Last Interglacial period: new U-series evidence from Hawaii and Bermuda and a new fossil compilation for North America. Quat Sci Rev 21:1355–1383
Nielsen R, Beaumont MA (2009) Statistical inferences in phylogeography. Mol Ecol 18:1034–1047
Otto-Bliesner BL, Marshall SJ, Overpeck JT, Miller GH, Hu A (2006) Simulating arctic climate warmth and icefield retreat in the last interglaciation. Science 311:1751–1753
Pellegrino K, Rodrigues MT, Waite AN, Morando M, Yonenaga-Yassuda Y, Sites JW (2005) Phylogeography and species limits in the Gymnodactylus darwinii complex (Gekkonidae, Squamata): genetic structure coincides with river systems in the Brazilian Atlantic Forest. Biol J Linn Soc Lond 85:13–26
Pessenda LCR, Oliveira PE, Mofatto M, Medeiros VB, Garcia RJF, Aravena R et al (2009) The evolution of a tropical rainforest/grassland mosaic in southeastern Brazil since 28,000 14 C yr BP based on carbon isotopes and pollen records. Quat Res 71:437–452
Porto TJ, Carnaval AC, Rocha PLB (2013) Evaluating forest refugial models using species distribution models, model filling and inclusion: a case study with 14 Brazilian species. Divers Distrib 19:330–340
Primmer CR, Borge T, Lindell J, Saetre GP (2002) Single nucleotide polymorphism characterization in species with limited available sequence information: high nucleotide diversity revealed in avian genome. Mol Ecol 11:603–612
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Core Team (2018) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Mol Biol Evol 19:2092–2100
Ribeiro RA, Lemos-Filho JP, Ramos ACS, Lovato MB (2010) Phylogeography of the endangered rosewood Dalbergia nigra (Fabaceae): insights into the evolutionary history and conservation of the Brazilian Atlantic Forest. Heredity 106:46–57
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor, Cold Spring Harbor Press, New York
Tajima F (1989) The effect of change in population size on DNA polymorphism. Genetics 123:597–601
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Terribile LC, Lima-Ribeiro MS, Araújo MB, Bizão N, Collevatti RG, Dobrovolski R et al (2012) Areas of climate stability of species ranges in the Brazilian Cerrado: disentangling uncertainties through time. Nat Conserv 10:152–159
Thomé MTC, Zamudio KR, Giovanelli JG, Haddad CF, Baldissera FA, Alexandrino J (2010) Phylogeography of endemic toads and post-Pliocene persistence of the Brazilian Atlantic Forest. Mol Phylogenet Evol 55:1018–1031
Thomé MTC, Zamudio KR, Haddad CF, Alexandrino J (2014) Barriers, rather than refugia, underlie the origin of diversity in toads endemic to the Brazilian Atlantic Forest. Mol Ecol 23:6152–6164
Thuiller W, Lafourcade B, Engler R, Araújo MB (2009) BIOMOD—a platform for ensemble forecasting of species distributions. Ecography 32:369–373
Tonini JFR, Costa LP, Carnaval AC (2013) Phylogeographic structure is strong in the Atlantic Forest; predictive power of correlative paleodistribution models, not always. J Zool Syst Evol Res 51:114–121
Valdez L, D’Elía G (2013) Differentiation in the Atlantic Forest: phylogeography of Akodon montensis (Rodentia, Sigmodontinae) and the Carnaval-Moritz model of Pleistocene refugia. J Mammal 94:911–922
Wallace AR (1854) On the monkeys of the Amazon. J Natl Hist 14:451–454
Weir JT, Schluter D (2008) Calibrating the avian molecular clock. Mol Ecol 17:2321–2328
Wisz MS, Pottier J, Kissling WD, Pellissier L, Lenoir J, Damgaard CF et al (2013) The role of biotic interactions in shaping distributions and realized assemblages of species: implications for species distribution modelling. Biol Rev Camb Philos Soc 88:3–15
Acknowledgements
The authors thank the Rede de Plataformas Tecnológicas for use of its sequencing facility in FIOCRUZ-Bahia, and the organizations which provided funds for this study: FAPESB (RED0045/2014; JCB0026/2016), CNPq (for scientific initiation grant and through project 443249/2014-8), CAPES, PROPCI/UFBA (PRODOC-2013/5813), FAPESP (2009/12989-1, BIOTA 2013/50297-0), NSF (DOB 1343578), and NASA. This work was developed in the Research Center on Biodiversity and Computing (BioComp) of the Universidade de São Paulo (USP), supported by the USP Provost’s Office for Research. We also thank the Laboratório de Biodiversidade e Evolução Molecular (LBEM) of the Universidade Federal de Minas Gerais, and Museu Nacional do Rio de Janeiro of the Universidade Federal do Rio de Janeiro for providing tissue samples for this study. We are grateful to Marcelo Vasconcelos for discussions and comments on previous versions of this manuscript. We thank two anonymous reviewers and the Editor Franz Bairlein for their comments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Data archiving
Sequence data have been submitted to GenBank: accession numbers cytb (MK610041-MK610097), G3PDH (MK671363-MK671418), TGFB2 (MK671419-MK671472).
Additional information
Communicated by J. T. Lifjeld.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mascarenhas, R., Miyaki, C.Y., Dobrovolski, R. et al. Late Pleistocene climate change shapes population divergence of an Atlantic Forest passerine: a model-based phylogeographic hypothesis test. J Ornithol 160, 733–748 (2019). https://doi.org/10.1007/s10336-019-01650-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10336-019-01650-1