Abstract
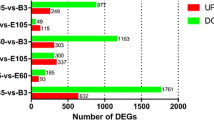
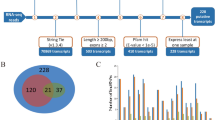
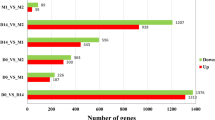
Myogenesis is accompanied by a number of changes in gene expression in mammals, and the transcriptional events that underlie these processes have not been yet fully elucidated. In this study, RNA-seq was used to comprehensively compare the transcription profiles of skeletal muscle between Tongcheng (TC) and Yorkshire (YK) pigs at 40, 55, 63, 70, and 90 days of gestation. One thousand three hundred seventeen and 691 differentially expressed genes (DEGs) were detected in TC and YK, respectively, among which 321 DEGs were shown to be common in TC and YK. STEM (Time-series Expression Miner) analysis revealed different gene expression profiles between the two breeds. One thousand six hundred seventy-seven genes showed significant differential expression between TC and YK at the identical stages, while three genes were found to be common in all comparisons. A total of 3185 new putative transcripts were also predicted. Several gene expression profiles were further validated by qRT-PCR. Fifty-five dpc (days post coitum) was suggested to be the key stage to contribute developmental differences between TC and YK. PTEN, EP300, ENSSSCG00000004979 (Myosin 9A), CDK14, IRS1, PPP1CC, and some ribosomal proteins were suggested to be the key candidate genes for elucidating the developmental differences between the two breeds. In conclusion, we constructed comprehensive high-resolution gene expression maps of these two pig breeds, which not only provides an in-depth understanding of the dynamics of transcriptional regulation during myogenesis in this study, but also would facilitate the elucidation of molecular mechanisms underlying myogenesis in the future studies.







Similar content being viewed by others
References
Anderson SJ, Lauritsen JP, Hartman MG, Foushee AM, Lefebvre JM, Shinton SA, Gerhardt B, Hardy RR, Oravecz T, Wiest DL (2007) Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity 26(6):759–772. https://doi.org/10.1016/j.immuni.2007.04.012*10.1016/j.immuni.2007.04.012
Cagnazzo M, Te PM, Priem J, de Wit AA, Pool MH, Davoli R, Russo V (2006) Comparison of prenatal muscle tissue expression profiles of two pig breeds differing in muscle characteristics. J Anim Sci 84(1):1–10. https://doi.org/10.2527/2006.8411
Chaillou T, Zhang X, McCarthy JJ (2016) Expression of muscle-specific ribosomal protein L3-like impairs myotube growth. J Cell Physiol 231(9):1894–1902. https://doi.org/10.1002/jcp.25294*10.1002/jcp.25294
Chalhoub N, Baker SJ (2009) PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol 4(1):127–150. https://doi.org/10.1146/annurev.pathol.4.110807.092311*10.1146/annurev.pathol.4.110807.092311
Chen J, Wang Y, Hamed M, Lacroix N, Li Q (2015) Molecular basis for the regulation of transcriptional coactivator p300 in myogenic differentiation. Sci Rep 5(1):13727. https://doi.org/10.1038/srep13727*10.1038/srep13727
Davoli R, Braglia S, Russo V, Varona L, Te PM (2011) Expression profiling of functional genes in prenatal skeletal muscle tissue in Duroc and Pietrain pigs. J Anim Breed Genet 128(1):15–27. https://doi.org/10.1111/j.1439-0388.2010.00867.x*10.1111/j.1439-0388.2010.00867.x
Deng H, Hershenson MB, Lei J, Bitar KN, Fingar DC, Solway J, Bentley JK (2010) p70 ribosomal S6 kinase is required for airway smooth muscle cell size enlargement but not increased contractile protein expression. Am J Respir Cell Mol Biol 42(6):744–752. https://doi.org/10.1165/rcmb.2009-0037OC*10.1165/rcmb.2009-0037OC
Djouder N, Metzler SC, Schmidt A, Wirbelauer C, Gstaiger M, Aebersold R, Hess D, Krek W (2007) S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Mol Cell 28(1):28–40. https://doi.org/10.1016/j.molcel.2007.08.010*10.1016/j.molcel.2007.08.010
Dunglison GF, Scotting PJ, Wigmore PM (1999) Rat embryonic myoblasts are restricted to forming primary fibres while later myogenic populations are pluripotent. Mech Dev 87(1-2):11–19. https://doi.org/10.1016/S0925-4773(99)00134-3
Ernst J, Bar-Joseph Z (2006) STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7(1):191. https://doi.org/10.1186/1471-2105-7-191*10.1186/1471-2105-7-191
Fan B, Tang Z, Xu S, Liu B, Peng Z, Li K (2006) Germplasm characteristics and conservation of Tongcheng pig: a case study for preservation and utilization of Chinese indigenous pig breeds. Anim Genet Resour Inf 39:51–63. https://doi.org/10.1017/S1014233900002133*10.1017/S1014233900002133
Florea L, Song L, Salzberg SL (2013) Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Res 2:188. https://doi.org/10.12688/f1000research.2-188.v2*10.12688/f1000research.2-188.v2
Ghandhi SA, Sinha A, Markatou M, Amundson SA (2011) Time-series clustering of gene expression in irradiated and bystander fibroblasts: an application of FBPA clustering. BMC Genomics 12(1):2. https://doi.org/10.1186/1471-2164-12-2*10.1186/1471-2164-12-2
Hao Y, Feng Y, Yang P, Cui Y, Liu J, Yang C, Gu X (2016) Transcriptome analysis reveals that constant heat stress modifies the metabolism and structure of the porcine longissimus dorsi skeletal muscle. Mol Gen Genomics 291(6):2101–2115. https://doi.org/10.1007/s00438-016-1242-8*10.1007/s00438-016-1242-8
Katsumata M (2011) Promotion of intramuscular fat accumulation in porcine muscle by nutritional regulation. Anim Sci J 82(1):17–25. https://doi.org/10.1111/j.1740-0929.2010.00844.x*10.1111/j.1740-0929.2010.00844.x
Kim NK, Lim JH, Song MJ, Kim OH, Park BY, Kim MJ, Hwang IH, Lee CS (2008) Comparisons of longissimus muscle metabolic enzymes and muscle fiber types in Korean and western pig breeds. Meat Sci 78(4):455–460. https://doi.org/10.1016/j.meatsci.2007.07.014*10.1016/j.meatsci.2007.07.014
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14(4):R36. https://doi.org/10.1186/gb-2013-14-4-r36*10.1186/gb-2013-14-4-r36
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10(3):R25. https://doi.org/10.1186/gb-2009-10-3-r25*10.1186/gb-2009-10-3-r25
Li RW, Schroeder SG (2012) Cytoskeleton remodeling and alterations in smooth muscle contractility in the bovine jejunum during nematode infection. Funct Integr Genomics 12(1):35–44. https://doi.org/10.1007/s10142-011-0259-7*10.1007/s10142-011-0259-7
Liu W, Thomas SG, Asa SL, Gonzalez-Cadavid N, Bhasin S, Ezzat S (2003) Myostatin is a skeletal muscle target of growth hormone anabolic action. J Clin Endocrinol Metab 88(11):5490–5496. https://doi.org/10.1210/jc.2003-030497*10.1210/jc.2003-030497
Long YC, Cheng Z, Copps KD, White MF (2011) Insulin receptor substrates Irs1 and Irs2 coordinate skeletal muscle growth and metabolism via the Akt and AMPK pathways. Mol Cell Biol 31(3):430–441. https://doi.org/10.1128/MCB.00983-10*10.1128/MCB.00983-10
Lunney JK (2007) Advances in swine biomedical model genomics. Int J Biol Sci 3(3):179–184
Ma X, Tang Z, Wang N, Zhao S, Wang R, Tan L, Mu Y, Li K (2011) Identification of extracellular matrix and cell adhesion molecule genes associated with muscle development in pigs. DNA Cell Biol 30(7):469–479. https://doi.org/10.1089/dna.2011.1218*10.1089/dna.2011.1218
Majewska M, Lipka A, Paukszto L, Jastrzebski JP, Myszczynski K, Gowkielewicz M, Jozwik M, Majewski MK (2017) Transcriptome profile of the human placenta. Funct Integr Genomics. 17(5):551–563. https://doi.org/10.1007/s10142-017-0555-y*10.1007/s10142-017-0555-y
McPherron AC, Lawler AM, Lee SJ (1997) Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387(6628):83–90. https://doi.org/10.1038/387083a0*10.1038/387083a0
Mirzayans R, Andrais B, Scott A, Murray D (2012) New insights into p53 signaling and cancer cell response to DNA damage: implications for cancer therapy. J Biomed Biotechnol 2012:170325. https://doi.org/10.1155/2012/170325*10.1155/2012/170325
Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G (1999) Drosophila S6 kinase: a regulator of cell size. Science 285(5436):2126–2129. https://doi.org/10.1126/science.285.5436.2126
Murani E, Muraniova M, Ponsuksili S, Schellander K, Wimmers K (2007) Identification of genes differentially expressed during prenatal development of skeletal muscle in two pig breeds differing in muscularity. BMC Dev Biol 7(1):109. https://doi.org/10.1186/1471-213X-7-109*10.1186/1471-213X-7-109
Nesteruk M, Hennig EE, Mikula M, Karczmarski J, Dzwonek A, Goryca K, Rubel T, Paziewska A, Woszczynski M, Ledwon J, Dabrowska M, Dadlez M, Ostrowski J (2014) Mitochondrial-related proteomic changes during obesity and fasting in mice are greater in the liver than skeletal muscles. Funct Integr Genomics 14(1):245–259. https://doi.org/10.1007/s10142-013-0342-3*10.1007/s10142-013-0342-3
Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40(12):1413–1415. https://doi.org/10.1038/ng.259*10.1038/ng.259
Perry RL, Rudnick MA (2000) Molecular mechanisms regulating myogenic determination and differentiation. Front Biosci 5(3):D750–D767. https://doi.org/10.2741/A548
Ponsuksili S, Murani E, Phatsara C, Schwerin M, Schellander K, Wimmers K (2009) Porcine muscle sensory attributes associate with major changes in gene networks involving CAPZB, ANKRD1, and CTBP2. Funct Integr Genomics 9(4):455–471. https://doi.org/10.1007/s10142-009-0131-1*10.1007/s10142-009-0131-1
Porrello A, Cerone MA, Coen S, Gurtner A, Fontemaggi G, Cimino L, Piaggio G, Sacchi A, Soddu S (2000) p53 regulates myogenesis by triggering the differentiation activity of pRb. J Cell Biol 151(6):1295–1304. https://doi.org/10.1083/jcb.151.6.1295
Porter HA, Perry A, Kingsley C, Tran NL, Keegan AD (2013) IRS1 is highly expressed in localized breast tumors and regulates the sensitivity of breast cancer cells to chemotherapy, while IRS2 is highly expressed in invasive breast tumors. Cancer Lett 338(2):239–248. https://doi.org/10.1016/j.canlet.2013.03.030*10.1016/j.canlet.2013.03.030
Qiu Z, Wan L, Chen T, Wan Y, He X, Lu S, Wang Y, Lin J (2013) The regulation of cambial activity in Chinese fir (Cunninghamia lanceolata) involves extensive transcriptome remodeling. New Phytol 199(3):708–719. https://doi.org/10.1111/nph.12301*10.1111/nph.12301
Ramocki NM, Wilkins HR, Magness ST, Simmons JG, Scull BP, Lee GH, McNaughton KK, Lund PK (2008) Insulin receptor substrate-1 deficiency promotes apoptosis in the putative intestinal crypt stem cell region, limits Apcmin/+ tumors, and regulates Sox9. Endocrinology 149(1):261–267. https://doi.org/10.1210/en.2007-0869*10.1210/en.2007-0869
Sabatini DM (2006) mTOR and cancer: insights into a complex relationship. Nat Rev Cancer 6(9):729–734. https://doi.org/10.1038/nrc1974*10.1038/nrc1974
Sebastian S, Faralli H, Yao Z, Rakopoulos P, Palii C, Cao Y, Singh K, Liu QC, Chu A, Aziz A, Brand M, Tapscott SJ, Dilworth FJ (2013) Tissue-specific splicing of a ubiquitously expressed transcription factor is essential for muscle differentiation. Genes Dev 27(11):1247–1259. https://doi.org/10.1101/gad.215400.113*10.1101/gad.215400.113
Sharples AP, Al-Shanti N, Hughes DC, Lewis MP, Stewart CE (2013) The role of insulin-like-growth factor binding protein 2 (IGFBP2) and phosphatase and tensin homologue (PTEN) in the regulation of myoblast differentiation and hypertrophy. Growth Hormon IGF Res 23:53–61. https://doi.org/10.1016/j.ghir.2013.03.004*10.1016/j.ghir.2013.03.004
Shin S, Song Y, Ahn J, Kim E, Chen P, Yang S, Suh Y, Lee K (2015) A novel mechanism of myostatin regulation by its alternative splicing variant during myogenesis in avian species. Am J Physiol Cell Physiol 309(10):C650–C659. https://doi.org/10.1152/ajpcell.00187.2015*10.1152/ajpcell.00187.2015
Shu F, Lv S, Qin Y, Ma X, Wang X, Peng X, Luo Y, Xu BE, Sun X, Wu J (2007) Functional characterization of human PFTK1 as a cyclin-dependent kinase. Proc Natl Acad Sci U S A 104(22):9248–9253. https://doi.org/10.1073/pnas.0703327104*10.1073/pnas.0703327104
Stadanlick JE, Zhang Z, Lee SY, Hemann M, Biery M, Carleton MO, Zambetti GP, Anderson SJ, Oravecz T, Wiest DL (2011) Developmental arrest of T cells in Rpl22-deficient mice is dependent upon multiple p53 effectors. J Immunol 187(2):664–675. https://doi.org/10.4049/jimmunol.1100029*10.4049/jimmunol.1100029
Sun K, Xue H, Wang H, Wang Q, Zuo XR, Xie WP, Wang H (2012) The effects of siRNA against RPL22 on ET-1-induced proliferation of human pulmonary arterial smooth muscle cells. Int J Mol Med 30(2):351–357. https://doi.org/10.3892/ijmm.2012.992*10.3892/ijmm.2012.992
Sun J, Xie M, Huang Z, Li H, Chen T, Sun R, Wang J, Xi Q, Wu T, Zhang Y (2017) Integrated analysis of non-coding RNA and mRNA expression profiles of 2 pig breeds differing in muscle traits. J Anim Sci 95(3):1092–1103. https://doi.org/10.2527/jas.2016.0867*10.2527/jas.2016.0867
Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39(Database):D561–D568. https://doi.org/10.1093/nar/gkq973*10.1093/nar/gkq973
Tang Z, Li Y, Wan P, Li X, Zhao S, Liu B, Fan B, Zhu M, Yu M, Li K (2007) LongSAGE analysis of skeletal muscle at three prenatal stages in Tongcheng and Landrace pigs. Genome Biol 8(6):R115. https://doi.org/10.1186/gb-2007-8-6-r115*10.1186/gb-2007-8-6-r115
Tang Z, Yang Y, Wang Z, Zhao S, Mu Y, Li K (2015) Integrated analysis of miRNA and mRNA paired expression profiling of prenatal skeletal muscle development in three genotype pigs. Sci Rep 5(1):15544. https://doi.org/10.1038/srep15544*10.1038/srep15544
Te PM, Hulsegge I, Coster A, Pool MH, Heuven HH, Janss LL (2007) Biochemical pathways analysis of microarray results: regulation of myogenesis in pigs. BMC Dev Biol 7:66. https://doi.org/10.1186/1471-213X-7-66*10.1186/1471-213X-7-66
Torok I, Herrmann-Horle D, Kiss I, Tick G, Speer G, Schmitt R, Mechler BM (1999) Down-regulation of RpS21, a putative translation initiation factor interacting with P40, produces viable minute imagos and larval lethality with overgrown hematopoietic organs and imaginal discs. Mol Cell Biol 19(3):2308–2321. https://doi.org/10.1128/MCB.19.3.2308
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515. https://doi.org/10.1038/nbt.1621*10.1038/nbt.1621
Valin C, Touraille C, Vigneron P, Ashmore CR (1982) Prediction of lamb meat quality traits based on muscle biopsy fibre typing. Meat Sci 6(4):257–263. https://doi.org/10.1016/0309-1740(82)90036-5*10.1016/0309-1740(82)90036-5
Vijayakumar A, Wu Y, Sun H, Li X, Jeddy Z, Liu C, Schwartz GJ, Yakar S, LeRoith D (2012) Targeted loss of GHR signaling in mouse skeletal muscle protects against high-fat diet-induced metabolic deterioration. Diabetes 61(1):94–103. https://doi.org/10.2337/db11-0814*10.2337/db11-0814
Vivanco I, Sawyers CL (2002) The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev Cancer 2(7):489–501. https://doi.org/10.1038/nrc839*10.1038/nrc839
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10(1):57–63. https://doi.org/10.1038/nrg2484*10.1038/nrg2484
Wang C, Gong B, Bushel PR, Thierry-Mieg J, Thierry-Mieg D, Xu J, Fang H, Hong H, Shen J, Su Z, Meehan J, Li X, Yang L, Li H, Labaj PP, Kreil DP, Megherbi D, Gaj S, Caiment F, van Delft J, Kleinjans J, Scherer A, Devanarayan V, Wang J, Yang Y, Qian HR, Lancashire LJ, Bessarabova M, Nikolsky Y, Furlanello C, Chierici M, Albanese D, Jurman G, Riccadonna S, Filosi M, Visintainer R, Zhang KK, Li J, Hsieh JH, Svoboda DL, Fuscoe JC, Deng Y, Shi L, Paules RS, Auerbach SS, Tong W (2014) The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat Biotechnol 32(9):926–932. https://doi.org/10.1038/nbt.3001*10.1038/nbt.3001
Wang Y, Zhao Y, Li J, Liu H, Ernst CW, Liu X, Liu G, Xi Y, Lei M (2015) Evaluation of housekeeping genes for normalizing real-time quantitative PCR assays in pig skeletal muscle at multiple developmental stages. Gene 565(2):235–241. https://doi.org/10.1016/j.gene.2015.04.016*10.1016/j.gene.2015.04.016
Wigmore PM, Stickland NC (1983) Muscle development in large and small pig fetuses. J Anat 137(Pt 2):235–245
Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124(3):471–484. https://doi.org/10.1016/j.cell.2006.01.016*10.1016/j.cell.2006.01.016
Xiong X, Zhao Y, He H, Sun Y (2011) Ribosomal protein S27-like and S27 interplay with p53-MDM2 axis as a target, a substrate and a regulator. Oncogene 30(15):1798–1811. https://doi.org/10.1038/onc.2010.569*10.1038/onc.2010.569
Xu YJ, Jin ML, Wang LJ, Zhang AD, Zuo B, Xu DQ, Ren ZQ, Lei MG, Mo XY, Li FE, Zheng R, Deng CY, Xiong YZ (2009) Differential proteome analysis of porcine skeletal muscles between Meishan and Large White. J Anim Sci 87(8):2519–2527. https://doi.org/10.2527/jas.2008-1708*10.2527/jas.2008-1708
Yang XR, Yu B, Mao XB, Zheng P, He J, Yu J, He Y, Reecy JM, Chen DW (2015) Lean and obese pig breeds exhibit differences in prenatal gene expression profiles of muscle development. Animal 9(01):28–34. https://doi.org/10.1017/S1751731114002316*10.1017/S1751731114002316
Yang Y, Tang Z, Fan X, Xu K, Mu Y, Zhou R, Li K (2016) Transcriptome analysis revealed chimeric RNAs, single nucleotide polymorphisms and allele-specific expression in porcine prenatal skeletal muscle. Sci Rep 6(1):29039. https://doi.org/10.1038/srep29039*10.1038/srep29039
Yao TP, SP O, Fuchs M, Zhou ND, Ch'Ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R (1998) Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93(3):361–372. https://doi.org/10.1016/S0092-8674(00)81165-4
Yu Y, Fuscoe JC, Zhao C, Guo C, Jia M, Qing T, Bannon DI, Lancashire L, Bao W, Du T, Luo H, Su Z, Jones WD, Moland CL, Branham WS, Qian F, Ning B, Li Y, Hong H, Guo L, Mei N, Shi T, Wang KY, Wolfinger RD, Nikolsky Y, Walker SJ, Duerksen-Hughes P, Mason CE, Tong W, Thierry-Mieg J, Thierry-Mieg D, Shi L, Wang C (2014) A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun 5:3230. https://doi.org/10.1038/ncomms4230*10.1038/ncomms4230
Yu Y, Li X, Liu L, Chai J, Haijun Z, Chu W, Yin H, Ma L, Duan H, Xiao M (2016) miR-628 promotes burn-induced skeletal muscle atrophy via targeting IRS1. Int J Biol Sci 12(10):1213–1224. https://doi.org/10.7150/ijbs.15496*10.7150/ijbs.15496
Zhan S, Zhao W, Song T, Dong Y, Guo J, Cao J, Zhong T, Wang L, Li L, Zhang H (2017) Dynamic transcriptomic analysis in hircine longissimus dorsi muscle from fetal to neonatal development stages. Funct Integr Genomics. https://doi.org/10.1007/s10142-017-0573-9*10.1007/s10142-017-0573-9
Zhang M, Zhu B, Davie J (2015) Alternative splicing of MEF2C pre-mRNA controls its activity in normal myogenesis and promotes tumorigenicity in rhabdomyosarcoma cells. J Biol Chem 290(1):310–324. https://doi.org/10.1074/jbc.M114.606277*10.1074/jbc.M114.606277
Zhang W, Liu R, Tang C, Xi Q, Lu S, Chen W, Zhu L, Cheng J, Chen Y, Wang W, Zhong J, Deng Y (2016) PFTK1 regulates cell proliferation, migration and invasion in epithelial ovarian cancer. Int J Biol Macromol 85:405–416. https://doi.org/10.1016/j.ijbiomac.2016.01.009*10.1016/j.ijbiomac.2016.01.009
Zhao X, Mo D, Li A, Gong W, Xiao S, Zhang Y, Qin L, Niu Y, Guo Y, Liu X, Cong P, He Z, Wang C, Li J, Chen Y (2011) Comparative analyses by sequencing of transcriptomes during skeletal muscle development between pig breeds differing in muscle growth rate and fatness. PLoS One 6(5):e19774. https://doi.org/10.1371/journal.pone.0019774*10.1371/journal.pone.0019774
Zhao H, Wang D, Fu D, Xue L (2015a) Predicting the potential ankylosing spondylitis-related genes utilizing bioinformatics approaches. Rheumatol Int 35(6):973–979. https://doi.org/10.1007/s00296-014-3178-9*10.1007/s00296-014-3178-9
Zhao Y, Li J, Liu H, Xi Y, Xue M, Liu W, Zhuang Z, Lei M (2015b) Dynamic transcriptome profiles of skeletal muscle tissue across 11 developmental stages for both Tongcheng and Yorkshire pigs. BMC Genomics 16(1):377. https://doi.org/10.1186/s12864-015-1580-7*10.1186/s12864-015-1580-7
Acknowledgments
We appreciate the Beijing Genomics Institute (BGI) and Gene Denovo for providing us with technical assistance in RNA sequencing and bioinformatics analysis; we also thank the help of Animal Husbandry Bureau of Tongcheng County.
Funding
This search project was supported financially by the National Porcine Industry Technology System (CARS-36).
Author information
Authors and Affiliations
Contributions
ML conceived this study and supervised the experiment. YZ and JL designed the breeding and sampling plan. HL, YX, GL, YZ, and JL participated in sampling. HL carried out the experiment and drafted the manuscript. HL analyzed the data. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Electronic supplementary material
The transcriptome raw data of Tongcheng and Yorkshire pigs were submitted to the Sequence Read Archive (SRA) of the NCBI (accession number: SRP 066398).
ESM Figure 1
Distribution of genes’ coverage (GIF 92 kb)
High Resolution Image
(TIFF 771 kb)
ESM Table 1
GO enrichment analysis of profile 39 in TC and that in YK (XLSX 309 kb)
ESM Table 2
GO enrichment analysis of profile 0 in TC and that in YK (XLSX 308 kb)
ESM Table 3
KEGG pathway analysis of the differentially expressed genes in specific profiles in TC and YK (XLSX 41 kb)
ESM Table 4
KEGG pathway analysis of the differentially expressed genes between TC and YK, and the genes involved in oxidative phosphorylation (XLSX 42 kb)
Table 5
The topological property of the networks with the DEGs between TC and YK (XLSX 120 kb)
ESM Table 6
The topological property of the networks with the stage-specific DEGs in 55 dpc between TC and YK (XLSX 48 kb)
ESM Table 7
The topological property of the networks with the up-regulated DEGs in TC at 55 dpc between TC and YK (XLSX 9 kb)
ESM Table 8
Novel transcripts predicted by RNA-seq (XLSX 914 kb)
ESM Table 9
KEGG pathway analysis of the genes which occurred alternative splicing events (XLSX 955 kb)
ESM Table 10
Alternative splicing events in each stage (XLSX 5462 kb)
Rights and permissions
About this article
Cite this article
Liu, H., Xi, Y., Liu, G. et al. Comparative transcriptomic analysis of skeletal muscle tissue during prenatal stages in Tongcheng and Yorkshire pig using RNA-seq. Funct Integr Genomics 18, 195–209 (2018). https://doi.org/10.1007/s10142-017-0584-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-017-0584-6