Abstract
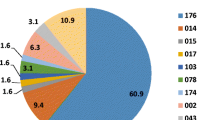
The molecular epidemiology of C. difficile strains causing disease in South Africa is currently unknown. Previously, multidrug resistant ribotype (RT)017 strains were those most commonly isolated from patients with diarrhoea attending Groote Schuur Hospital in Cape Town, South Africa. This larger study aimed to investigate the molecular epidemiology and antibiotic susceptibility profiles of C. difficile strains in the greater Cape Town and regional areas. C. difficile strains were isolated from patients with diarrhoea attending hospitals in the Western Cape region of South Africa that tested positive using the GeneXpert CDiff diagnostic test. Ribotyping and multilocus variable-number tandem-repeat analysis (MLVA) were used to type isolates, and their susceptibilities to several antibiotics were determined by gradient diffusion test strips. A total of 269 non-repeat C. difficile isolates were obtained. A large proportion of isolates (64.3 %) belonged to the RT017 group, many of which were clonally related when investigated by MLVA. RT017 strains were particularly prevalent in patients attending specialist tuberculosis (TB) hospitals. The majority of RT017 isolates were co-resistant to moxifloxacin and rifampicin, two antibiotics which are used intensively during anti-TB therapy. Non-RT017 strains were generally susceptible to both antibiotics. Resistance to erythromycin was observed for both groups of strains. RT017 C. difficile strains are the most commonly isolated strains from patients attending healthcare facilities in the greater Cape Town and regional areas. The presence of multidrug resistant RT017 strains in patients with diarrhoea attending local TB hospitals reflects a potential reservoir for future infections.




Similar content being viewed by others
References
Levy AR, Szabo SM, Lozano-Ortega G, Lloyd-Smith E, Leung V, Lawrence R, Romney MG (2015) Incidence and costs of Clostridium difficile infections in Canada. Open Forum Infect Dis 2:ofv076. doi:10.1093/ofid/ofv076
Gabriel L, Beriot-Mathiot A (2014) Hospitalization stay and costs attributable to Clostridium difficile infection: a critical review. J Hosp Infect 88:12–21. doi:10.1016/j.jhin.2014.04.011
Wiegand PN, Nathwani D, Wilcox MH, Stephens J, Shelbaya A, Haider S (2012) Clinical and economic burden of Clostridium difficile infection in Europe: a systematic review of healthcare-facility-acquired infection. J Hosp Infect 81:1–14. doi:10.1016/j.jhin.2012.02.004
Cowardin CA, Buonomo EL, Saleh MM, Wilson MG, Burgess SL, Kuehne SA, Schwan C, Eichhoff AM, Koch-Nolte F, Lyras D, Aktories K, Minton NP, Petri WA (2016) The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia. Nat Microbiol 1:16108. doi:10.1038/nmicrobiol.2016.108
Chitnis AS, Holzbauer SM, Belflower RM, Winston LG, Bamberg WM, Lyons C, Farley MM, Dumyati GK, Wilson LE, Beldavs ZG, Dunn JR, Gould LH, MacCannell DR, Gerding DN, McDonald LC, Lessa FC (2013) Epidemiology of community-associated Clostridium difficile infection, 2009 through 2011. JAMA Intern Med 173:1359–1367. doi:10.1001/jamainternmed.2013.7056
Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC (2015) Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi:10.1056/NEJMoa1408913
McDonald LC, Killgore GE, Thompson A, Owens RC, Kazakova SV, Sambol SP, Johnson S, Gerding DN (2005) An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med 353:2433–2441. doi:10.1056/NEJMoa051590
Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC (2005) Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084. doi:10.1016/S0140-6736(05)67420-X
Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault A-M, Nguyen T, Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson TJ, Horn R, René P, Monczak Y, Dascal A (2005) A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med 353:2442–2449. doi:10.1056/NEJMoa051639
Bauer MP, Notermans DW, van Benthem BHB, Brazier JS, Wilcox MH, Rupnik M, Monnet DL, van Dissel JT, Kuijper EJ, ECDIS Study Group (2011) Clostridium difficile infection in Europe: a hospital-based survey. Lancet 377:63–73. doi:10.1016/S0140-6736(10)61266-4
Freeman J, Vernon J, Morris K, Nicholson S, Todhunter S, Longshaw C, Wilcox MH, Pan-European Longitudinal Surveillance of Antibiotic Resistance among Prevalent Clostridium difficile Ribotypes’ Study Group (2015) Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect 21:248.e9–248.e16. doi:10.1016/j.cmi.2014.09.017
Hawkey PM, Marriott C, Liu WE, Jian ZJ, Gao Q, Ling TKW, Chow V, So E, Chan R, Hardy K, Xu L, Manzoor S (2013) Molecular epidemiology of Clostridium difficile infection in a major Chinese hospital: an underrecognized problem in Asia? J Clin Microbiol 51:3308–3313. doi:10.1128/JCM.00587-13
Huang H, Weintraub A, Fang H, Wu S, Zhang Y, Nord CE (2010) Antimicrobial susceptibility and heteroresistance in Chinese Clostridium difficile strains. Anaerobe 16:633–635. doi:10.1016/j.anaerobe.2010.09.002
Lee J-H, Lee Y, Lee K, Riley TV, Kim H (2014) The changes of PCR ribotype and antimicrobial resistance of Clostridium difficile in a tertiary care hospital over 10 years. J Med Microbiol 63:819–823. doi:10.1099/jmm.0.072082-0
Kim H, Jeong SH, Roh KH, Hong SG, Kim JW, Shin M-G, Kim M-N, Shin HB, Uh Y, Lee H, Lee K (2010) Investigation of toxin gene diversity, molecular epidemiology, and antimicrobial resistance of Clostridium difficile isolated from 12 hospitals in South Korea. Korean J Lab Med 30:491–497. doi:10.3343/kjlm.2010.30.5.491
Ngamskulrungroj P, Sanmee S, Putsathit P, Piewngam P, Elliott B, Riley TV, Kiratisin P (2015) Molecular epidemiology of Clostridium difficile infection in a large teaching hospital in Thailand. PLoS One 10:e0127026. doi:10.1371/journal.pone.0127026
Drudy D, Harnedy N, Fanning S, Hannan M, Kyne L (2007) Emergence and control of fluoroquinolone-resistant, toxin A-negative, toxin B-positive Clostridium difficile. Infect Control Hosp Epidemiol 28:932–940. doi:10.1086/519181
Drudy D, Quinn T, O’Mahony R, Kyne L, O’Gaora P, Fanning S (2006) High-level resistance to moxifloxacin and gatifloxacin associated with a novel mutation in gyrB in toxin-A-negative, toxin-B-positive Clostridium difficile. J Antimicrob Chemother 58:1264–1267. doi:10.1093/jac/dkl398
Cairns MD, Preston MD, Lawley TD, Clark TG, Stabler RA, Wren BW (2015) Genomic epidemiology of a protracted hospital outbreak caused by a toxin A-negative Clostridium difficile sublineage PCR ribotype 017 strain in London, England. J Clin Microbiol 53:3141–3147. doi:10.1128/JCM.00648-15
Goorhuis A, Legaria MC, van den Berg RJ, Harmanus C, Klaassen CHW, Brazier JS, Lumelsky G, Kuijper EJ (2009) Application of multiple-locus variable-number tandem-repeat analysis to determine clonal spread of toxin A-negative Clostridium difficile in a general hospital in Buenos Aires, Argentina. Clin Microbiol Infect 15:1080–1086. doi:10.1111/j.1469-0691.2009.02759.x
Brown KA, Khanafer N, Daneman N, Fisman DN (2013) Meta-analysis of antibiotics and the risk of community-associated Clostridium difficile infection. Antimicrob Agents Chemother 57:2326–2332. doi:10.1128/AAC.02176-12
Slimings C, Riley TV (2014) Antibiotics and hospital-acquired Clostridium difficile infection: update of systematic review and meta-analysis. J Antimicrob Chemother 69:881–891. doi:10.1093/jac/dkt477
Vardakas KZ, Trigkidis KK, Boukouvala E, Falagas ME (2016) Clostridium difficile infection following systemic antibiotic administration in randomised controlled trials: a systematic review and meta-analysis. Int J Antimicrob Agents 48:1–10. doi:10.1016/j.ijantimicag.2016.03.008
Obuch-Woszczatyński P, Dubiel G, Harmanus C, Kuijper E, Duda U, Wultańska D, van Belkum A, Pituch H (2013) Emergence of Clostridium difficile infection in tuberculosis patients due to a highly rifampicin-resistant PCR ribotype 046 clone in Poland. Eur J Clin Microbiol Infect Dis 32:1027–1030. doi:10.1007/s10096-013-1845-5
Chang KC, Leung CC, Yew WW, Lam FM, Ho PL, Chau CH, Cheng VCC, Yuen KY (2009) Analyses of fluoroquinolones and Clostridium difficile-associated diarrhoea in tuberculosis patients. Int J Tuberc Lung Dis 13:341–346
Lee YM, Huh KC, Yoon SM, Jang BI, Shin JE, Koo HS, Jung Y, Kim SH, Moon HS, Lee SW, Daejeon-Chungchung Intestinal Research Group (2016) Incidence and clinical outcomes of Clostridium difficile infection after treatment with tuberculosis medication. Gut Liver 10:250–254. doi:10.5009/gnl14435
Sun Y-X, Zhao Y-T, Teng L-L, Ge J-L, Jiang H, Shao L (2013) Clostridium difficile infection associated with antituberculous agents in a patient with tuberculous pericarditis. Intern Med Tokyo Jpn 52:1495–1497. doi:10.2169/internalmedicine.52.0162
Jung S-W, Jeon S-W, Do B-H, Kim S-G, Ha S-S, Cho C-M, Tak W-Y, Kweon Y-O, Kim S-K, Choi Y-H, Cha S-I (2007) Clinical aspects of rifampicin-associated pseudomembranous colitis. J Clin Gastroenterol 41:38–40. doi:10.1097/MCG.0b013e31802dfaf7
Wang P, Zhou Y, Wang Z, Xie S, Zhang T, Lin M, Li R, Tan J, Chen Y, Jiang B (2014) Identification of Clostridium difficile ribotype 027 for the first time in Mainland China. Infect Control Hosp Epidemiol 35:95–98. doi:10.1086/674405
Kullin B, Meggersee R, D’Alton J, Galvão B, Rajabally N, Whitelaw A, Bamford C, Reid SJ, Abratt VR (2015) Prevalence of gastrointestinal pathogenic bacteria in patients with diarrhoea attending Groote Schuur Hospital, Cape Town, South Africa. South Afr Med J Suid-Afr Tydskr Vir Geneeskd 105:121–125. doi:10.7196/samj.8654
Lekalakala MR, Lewis E, Hoosen AA (2010) Clostridium difficile infections in a tertiary hospital: value of surveillance. J Hosp Infect 75:328–329. doi:10.1016/j.jhin.2010.03.016
Samie A, Obi CL, Franasiak J, Archbald-Pannone L, Bessong PO, Alcantara-Warren C, Guerrant RL (2008) PCR detection of Clostridium difficile triose phosphate isomerase (tpi), toxin A (tcdA), toxin B (tcdB), binary toxin (cdtA, cdtB), and tcdC genes in Vhembe District, South Africa. Am J Trop Med Hyg 78:577–585
Kullin B, Brock T, Rajabally N, Anwar F, Vedantam G, Reid S, Abratt V (2016) Characterisation of Clostridium difficile strains isolated from Groote Schuur Hospital, Cape Town, South Africa. Eur J Clin Microbiol Infect Dis. 35:1709–1718. doi:10.1007/s10096-016-2717-6
Brazier JS (1993) Role of the laboratory in investigations of Clostridium difficile diarrhea. Clin Infect Dis 16(Suppl 4):S228–S233. doi:10.1093/clinids/16.supplement_4.s228
Rajabally N, Kullin B, Ebrahim K, Brock T, Weintraub A, Whitelaw A, Bamford C, Watermeyer G, Thomson S, Abratt V, Reid S (2016) A comparison of Clostridium difficile diagnostic methods for identification of local strains in a South African centre. J Med Microbiol. doi:10.1099/jmm.0.000231
Lemee L, Dhalluin A, Testelin S, Mattrat M-A, Maillard K, Lemeland J-F, Pons J-L (2004) Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (Toxin A), and tcdB (Toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol 42:5710–5714. doi:10.1128/JCM.42.12.5710-5714.2004
Stubbs S, Rupnik M, Gibert M, Brazier J, Duerden B, Popoff M (2000) Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol Lett 186:307–312. doi:10.1016/S0378-1097(00)00162-2
Bidet P, Barbut F, Lalande V, Burghoffer B, Petit JC (1999) Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol Lett 175:261–266. doi:10.1111/j.1574-6968.1999.tb13629.x
Indra A, Huhulescu S, Schneeweis M, Hasenberger P, Kernbichler S, Fiedler A, Wewalka G, Allerberger F, Kuijper EJ (2008) Characterization of Clostridium difficile isolates using capillary gel electrophoresis-based PCR ribotyping. J Med Microbiol 57:1377–1382. doi:10.1099/jmm.0.47714-0
Martinson JNV, Broadaway S, Lohman E, Johnson C, Alam MJ, Khaleduzzaman M, Garey KW, Schlackman J, Young VB, Santhosh K, Rao K, Lyons RH, Walk ST (2015) Evaluation of portability and cost of a fluorescent PCR ribotyping protocol for Clostridium difficile epidemiology. J Clin Microbiol 53:1192–1197. doi:10.1128/JCM.03591-14
van den Berg RJ, Schaap I, Templeton KE, Klaassen CHW, Kuijper EJ (2007) Typing and subtyping of Clostridium difficile isolates by using multiple-locus variable-number tandem-repeat analysis. J Clin Microbiol 45:1024–1028. doi:10.1128/JCM.02023-06
Salipante SJ, Hall BG (2011) Inadequacies of minimum spanning trees in molecular epidemiology. J Clin Microbiol 49:3568–3575. doi:10.1128/JCM.00919-11
Claro T, Daniels S, Humphreys H (2014) Detecting Clostridium difficile spores from inanimate surfaces of the hospital environment: which method is best? J Clin Microbiol 52:3426–3428. doi:10.1128/JCM.01011-14
Ali S, Muzslay M, Wilson P (2015) A novel quantitative sampling technique for detection and monitoring of Clostridium difficile contamination in the clinical environment. J Clin Microbiol 53:2570–2574. doi:10.1128/JCM.00376-15
Faires MC, Pearl DL, Berke O, Reid-Smith RJ, Weese JS (2013) The identification and epidemiology of meticillin-resistant Staphylococcus aureus and Clostridium difficile in patient rooms and the ward environment. BMC Infect Dis 13:342. doi:10.1186/1471-2334-13-342
Verity P, Wilcox MH, Fawley W, Parnell P (2001) Prospective evaluation of environmental contamination by Clostridium difficile in isolation side rooms. J Hosp Infect 49:204–209. doi:10.1053/jhin.2001.1078
Dumford DM, Nerandzic MM, Eckstein BC, Donskey CJ (2009) What is on that keyboard? Detecting hidden environmental reservoirs of Clostridium difficile during an outbreak associated with North American pulsed-field gel electrophoresis type 1 strains. Am J Infect Control 37:15–19. doi:10.1016/j.ajic.2008.07.009
Clinical and Laboratory Standards Institute (2014) Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. CLSI document M100-S24, 1st edn. Clinical and Laboratory Standards Institute, Wayne
EUCAST (2016) The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 6.0, 2016 (http://www.eucast.org). Accessed 22 September 2016
Baines SD, O’Connor R, Freeman J, Fawley WN, Harmanus C, Mastrantonio P, Kuijper EJ, Wilcox MH (2008) Emergence of reduced susceptibility to metronidazole in Clostridium difficile. J Antimicrob Chemother 62:1046–1052. doi:10.1093/jac/dkn313
Rajabally NM, Pentecost M, Pretorius G, Whitelaw A, Mendelson M, Watermeyer G (2013) The Clostridium difficile problem: a South African tertiary institution’s prospective perspective. South Afr Med J Suid-Afr Tydskr Vir Geneeskd 103:168–172. doi:10.7196/samj.6012
Monot M, Eckert C, Lemire A, Hamiot A, Dubois T, Tessier C, Dumoulard B, Hamel B, Petit A, Lalande V, Ma L, Bouchier C, Barbut F, Dupuy B (2015) Clostridium difficile: new insights into the evolution of the pathogenicity locus. Sci Rep 5:15023. doi:10.1038/srep15023
Geric Stare B, Rupnik M (2010) Clostridium difficile toxinotype XI (A-B-) exhibits unique arrangement of PaLoc and its upstream region. Anaerobe 16:393–395. doi:10.1016/j.anaerobe.2010.04.001
Kim SJ, Kim H, Seo Y, Yong D, Jeong SH, Chong Y, Lee K (2010) Molecular characterization of toxin A-negative, toxin B-positive variant strains of Clostridium difficile isolated in Korea. Diagn Microbiol Infect Dis 67:198–201. doi:10.1016/j.diagmicrobio.2010.01.007
Landelle C, Verachten M, Legrand P, Girou E, Barbut F, Brun-Buisson C, Buisson CB (2014) Contamination of healthcare workers’ hands with Clostridium difficile spores after caring for patients with C. difficile infection. Infect Control Hosp Epidemiol 35:10–15. doi:10.1086/674396
Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, Kuijper EJ, Wilcox MH (2010) The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev 23:529–549. doi:10.1128/CMR.00082-09
Vuotto C, Moura I, Barbanti F, Donelli G, Spigaglia P (2016) Subinhibitory concentrations of metronidazole increase biofilm formation in Clostridium difficile strains. Pathog Dis 74. doi:10.1093/femspd/ftv114
Vardakas KZ, Polyzos KA, Patouni K, Rafailidis PI, Samonis G, Falagas ME (2012) Treatment failure and recurrence of Clostridium difficile infection following treatment with vancomycin or metronidazole: a systematic review of the evidence. Int J Antimicrob Agents 40:1–8. doi:10.1016/j.ijantimicag.2012.01.004
National Health Laboratory Service (2013) Western Cape academic hospitals antimicrobial recommendations [Internet]. http://www.fidssa.co.za/Content/Documents/Antibiotic_Rec_2013_final.pdf. Accessed 22 September 2016
Boyles TH, Whitelaw A, Bamford C, Moodley M, Bonorchis K, Morris V, Rawoot N, Naicker V, Lusakiewicz I, Black J, Stead D, Lesosky M, Raubenheimer P, Dlamini S, Mendelson M (2013) Antibiotic stewardship ward rounds and a dedicated prescription chart reduce antibiotic consumption and pharmacy costs without affecting inpatient mortality or re-admission rates. PLoS ONE 8. doi:10.1371/journal.pone.0079747
Johnson S, Schriever C, Galang M, Kelly CP, Gerding DN (2007) Interruption of recurrent Clostridium difficile-associated diarrhea episodes by serial therapy with vancomycin and rifaximin. Clin Infect Dis 44:846–848. doi:10.1086/511870
O’Connor JR, Galang MA, Sambol SP, Hecht DW, Vedantam G, Gerding DN, Johnson S (2008) Rifampin and rifaximin resistance in clinical isolates of Clostridium difficile. Antimicrob Agents Chemother 52:2813–2817. doi:10.1128/AAC.00342-08
Miller MA, Blanchette R, Spigaglia P, Barbanti F, Mastrantonio P (2011) Divergent rifamycin susceptibilities of Clostridium difficile strains in Canada and Italy and predictive accuracy of rifampin Etest for rifamycin resistance. J Clin Microbiol 49:4319–4321. doi:10.1128/JCM.05100-11
Curry SR, Marsh JW, Shutt KA, Muto CA, O’Leary MM, Saul MI, Pasculle AW, Harrison LH (2009) High frequency of rifampin resistance identified in an epidemic Clostridium difficile clone from a large teaching hospital. Clin Infect Dis 48:425–429. doi:10.1086/596315
Polage CR, Gyorke CE, Kennedy MA, Leslie JL, Chin DL, Wang S, Nguyen HH, Huang B, Tang Y-W, Lee LW, Kim K, Taylor S, Romano PS, Panacek EA, Goodell PB, Solnick JV, Cohen SH (2015) Overdiagnosis of Clostridium difficile infection in the molecular test era. JAMA Intern Med 1–10. doi:10.1001/jamainternmed.2015.4114
Acknowledgments
The authors would like to thank the staff at the NHLS Microbiology laboratory at GSH for initial processing of the samples, Dr Diane Rip & Mrs Zubeida Salaam-Dreyer for assistance with sample plating, Mrs Elzane Cronje and the NHLS media lab for preparing the CCEY agar plates, the IC staff at GSH for environmental sampling, Ms Alvera Vorster for assistance with the capillary gel electrophoresis experiments, Ms Marilyn Krige for assistance with some of the antibiotic gradient strip experiments and Liofilchem (BD diagnostics) for the donation of some rifampicin gradient strips.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by the National Research Funding Foundation of South Africa and the South African Medical Research Council. B. Kullin acknowledges the Claude Leon Foundation and the Carnegie Corporation of New York for Postdoctoral Fellowships.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committees and with the Helsinki declaration and its later amendments or comparable ethical standards. This study was approved by the Ethics Committee of the University of Cape Town (HREC Number: 310/2008). For this type of observational and retrospective study formal consent is not required.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
Minimum spanning tree representation of MLVA data for C. difficile RT017 isolates. Unique MLVA types are represented by individual circles, coloured according to the hospital the patient was in at the time of sample submission. The size of the circle represents the number of isolates per MLVA type and the circle labels indicate isolation date (year/month/date). Numbers between the circles represent the summed tandem repeats (STRDs) between isolates. Isolates with a STRD ≤ 2 are shaded to form clonally related groups (labelled by large numbers 1-7). The tree has been redrawn for ease of viewing and is not to scale. (PDF 887 kb)
Rights and permissions
About this article
Cite this article
Kullin, B., Wojno, J., Abratt, V. et al. Toxin A-negative toxin B-positive ribotype 017 Clostridium difficile is the dominant strain type in patients with diarrhoea attending tuberculosis hospitals in Cape Town, South Africa. Eur J Clin Microbiol Infect Dis 36, 163–175 (2017). https://doi.org/10.1007/s10096-016-2790-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-016-2790-x