Abstract
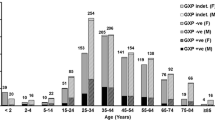
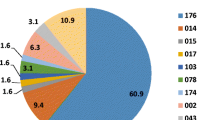
The C. difficile infection rate in South Africa is concerning. Many strains previously isolated from diarrhetic patients at Groote Schuur Hospital were ribotype 017. This study further characterised these strains with respect to their clonal relationships, antibiotic susceptibility, toxin production and various attributes impacting on pathogen colonisation. Multilocus variable-number tandem-repeat analysis (MLVA) was used to characterise all C. difficile isolates. Antibiotic susceptibility was determined by E-test and PCR-based analysis of the ermB, gyrA and gyrB genes. Auto-aggregation of cells was measured in broth, and biofilm formation observed in 24-well plates. Toxins were measured using the Wampole C DIFF TOX A/B II kit. Most isolates belonged to the ribotype 017 group. Identical MLVA types occurred in different wards over time, and several patients were infected with identical strains. All isolates were susceptible to vancomycin and metronidazole, but some ribotype 017 isolates showed reduced metronidazole susceptibility (≥2 mg l−1). Sixty-nine percent of ribotype 017 isolates were resistant to moxifloxacin, and 94 % to erythromycin, compared to 0 % and 17 % resistance, respectively, in non-ribotype 017 isolates. The ermB gene and mutations in the gyrA and/or gyrB genes were linked to erythromycin and moxifloxacin resistance, respectively. Ribotype 017 isolates auto-aggregated more strongly than other isolates and produced lower levels of the TcdB toxin than a reference strain. Certain strains produced strong biofilms. Patient-to-patient transfer and unique infection events could cause the predominance of ribotype 017 strains in the cohort. Multi-drug resistant strains are a potential reservoir for future infections.




Similar content being viewed by others
References
Levy AR, Szabo SM, Lozano-Ortega G, Lloyd-Smith E, Leung V, Lawrence R, Romney MG (2015) Incidence and costs of Clostridium difficile infections in Canada. Open Forum Infect Dis 2:ofv076. doi:10.1093/ofid/ofv076
Wiegand PN, Nathwani D, Wilcox MH, Stephens J, Shelbaya A, Haider S (2012) Clinical and economic burden of Clostridium difficile infection in Europe: a systematic review of healthcare-facility-acquired infection. J Hosp Infect 81:1–14. doi:10.1016/j.jhin.2012.02.004
Gabriel L, Beriot-Mathiot A (2014) Hospitalization stay and costs attributable to Clostridium difficile infection: a critical review. J Hosp Infect 88:12–21. doi:10.1016/j.jhin.2014.04.011
Chitnis AS, Holzbauer SM, Belflower RM, Winston LG, Bamberg WM, Lyons C, Farley MM, Dumyati GK, Wilson LE, Beldavs ZG, Dunn JR, Gould LH, MacCannell DR, Gerding DN, McDonald LC, Lessa FC (2013) Epidemiology of community-associated Clostridium difficile infection, 2009 through 2011. JAMA Int Med 173:1359–67. doi:10.1001/jamainternmed.2013.7056
Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC (2015) Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–34. doi:10.1056/NEJMoa1408913
Aguayo C, Flores R, Lévesque S, Araya P, Ulloa S, Lagos J, Hormazabal JC, Tognarelli J, Ibáñez D, Pidal P, Duery O, Olivares B, Fernández J (2015) Rapid spread of Clostridium difficile NAP1/027/ST1 in Chile confirms the emergence of the epidemic strain in Latin America. Epidemiol Infect 143:3069–73. doi:10.1017/S0950268815000023
Quesada-Gómez C, Rodríguez C, Gamboa-Coronado M del M, Rodríguez-Cavallini E, Du T, Mulvey MR, Villalobos-Zúñiga M, Boza-Cordero R (2010) Emergence of Clostridium difficile NAP1 in Latin America. J Clin Microbiol 48:669–70. doi:10.1128/JCM.02196-09
Bauer MP, Notermans DW, van Benthem BHB, Brazier JS, Wilcox MH, Rupnik M, Monnet DL, van Dissel JT, Kuijper EJ, ECDIS Study Group (2011) Clostridium difficile infection in Europe: a hospital-based survey. Lancet 377:63–73. doi:10.1016/S0140-6736(10)61266-4
Freeman J, Vernon J, Morris K, Nicholson S, Todhunter S, Longshaw C, Wilcox MH, Pan-European Longitudinal Surveillance of Antibiotic Resistance among Prevalent Clostridium difficile Ribotypes’ Study Group (2015) Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect Dis. 21:248.e9-248.e16. doi: 10.1016/j.cmi.2014.09.017
Hawkey PM, Marriott C, Liu WE, Jian ZJ, Gao Q, Ling TKW, Chow V, So E, Chan R, Hardy K, Xu L, Manzoor S (2013) Molecular epidemiology of Clostridium difficile infection in a major Chinese hospital: an underrecognized problem in Asia? J Clin Microbiol 51:3308–13. doi:10.1128/JCM.00587-13
Huang H, Weintraub A, Fang H, Wu S, Zhang Y, Nord CE (2010) Antimicrobial susceptibility and heteroresistance in Chinese Clostridium difficile strains. Anaerobe 16:633–5. doi:10.1016/j.anaerobe.2010.09.002
Lee J-H, Lee Y, Lee K, Riley TV, Kim H (2014) The changes of PCR ribotype and antimicrobial resistance of Clostridium difficile in a tertiary care hospital over 10 years. J Med Microbiol 63:819–23. doi:10.1099/jmm.0.072082-0
Kim H, Jeong SH, Roh KH, Hong SG, Kim JW, Shin M-G, Kim M-N, Shin HB, Uh Y, Lee H, Lee K (2010) Investigation of toxin gene diversity, molecular epidemiology, and antimicrobial resistance of Clostridium difficile isolated from 12 hospitals in South Korea. Korean J Lab Med 30:491–7. doi:10.3343/kjlm.2010.30.5.491
Ngamskulrungroj P, Sanmee S, Putsathit P, Piewngam P, Elliott B, Riley TV, Kiratisin P (2015) Molecular epidemiology of Clostridium difficile infection in a large teaching hospital in Thailand. PLoS One 10:e0127026. doi:10.1371/journal.pone.0127026
Drudy D, Harnedy N, Fanning S, Hannan M, Kyne L (2007) Emergence and control of fluoroquinolone-resistant, toxin A-negative, toxin B-positive Clostridium difficile. Infect Control Hosp Epidemiol 28:932–40. doi:10.1086/519181
Shin B-M, Kuak EY, Yoo SJ, Shin WC, Yoo HM (2008) Emerging toxin A-B+ variant strain of Clostridium difficile responsible for pseudomembranous colitis at a tertiary care hospital in Korea. Diagn Microbiol Infect Dis 60:333–7. doi:10.1016/j.diagmicrobio.2007.10.022
Cairns MD, Preston MD, Lawley TD, Clark TG, Stabler RA, Wren BW (2015) Genomic epidemiology of a protracted hospital outbreak caused by a toxin A-negative Clostridium difficile sublineage PCR ribotype 017 strain in London. Engl J Clin Microbiol 53:3141–7. doi:10.1128/JCM.00648-15
Goorhuis A, Legaria MC, van den Berg RJ, Harmanus C, Klaassen CHW, Brazier JS, Lumelsky G, Kuijper EJ (2009) Application of multiple-locus variable-number tandem-repeat analysis to determine clonal spread of toxin A-negative Clostridium difficile in a general hospital in Buenos Aires. Argent Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis 15:1080–6. doi:10.1111/j.1469-0691.2009.02759.x
Drudy D, Quinn T, O’Mahony R, Kyne L, O’Gaora P, Fanning S (2006) High-level resistance to moxifloxacin and gatifloxacin associated with a novel mutation in gyrB in toxin-A-negative, toxin-B-positive Clostridium difficile. J Antimicrob Chemother 58:1264–7. doi:10.1093/jac/dkl398
Kullin B, Meggersee R, D’Alton J, Galvão B, Rajabally N, Whitelaw A, Bamford C, Reid SJ, Abratt VR (2015) Prevalence of gastrointestinal pathogenic bacteria in patients with diarrhoea attending Groote Schuur Hospital, Cape Town. S Afr Med J 105:121–5. doi:10.7196/samj.8654
Lekalakala MR, Lewis E, Hoosen AA (2010) Clostridium difficile infections in a tertiary hospital: value of surveillance. J Hosp Infect 75:328–9. doi:10.1016/j.jhin.2010.03.016
Rajabally NM, Pentecost M, Pretorius G, Whitelaw A, Mendelson M, Watermeyer G (2013) The Clostridium difficile problem: a South African tertiary institution’s prospective perspective. S Afr Med J 103:168–72. doi:10.7196/samj.6012
Samie A, Obi CL, Franasiak J, Archbald-Pannone L, Bessong PO, Alcantara-Warren C, Guerrant RL (2008) PCR detection of Clostridium difficile triose phosphate isomerase (tpi), toxin A (tcdA), toxin B (tcdB), binary toxin (cdtA, cdtB), and tcdC genes in Vhembe District, South Africa. Am J Trop Med Hyg 78:577–85
Rajabally N, Kullin B, Ebrahim K, Brock T, Weintraub A, Whitelaw A, Bamford C, Watermeyer G, Thomson S, Abratt V, Reid S (2016) A comparison of Clostridium difficile diagnostic methods for identification of local strains in a South African centre. J Med Microbiol. doi:10.1099/jmm.0.000231
Eckert C, Vromman F, Halkovich A, Barbut F (2011) Multilocus variable-number tandem repeat analysis: a helpful tool for subtyping French Clostridium difficile PCR ribotype 027 isolates. J Med Microbiol 60:1088–94. doi:10.1099/jmm.0.029009-0
Marsh JW, O’Leary MM, Shutt KA, Pasculle AW, Johnson S, Gerding DN, Muto CA, Harrison LH (2006) Multilocus variable-number tandem-repeat analysis for investigation of Clostridium difficile transmission in hospitals. J Clin Microbiol 44:2558–66. doi:10.1128/JCM.02364-05
van den Berg RJ, Schaap I, Templeton KE, Klaassen CHW, Kuijper EJ (2007) Typing and subtyping of Clostridium difficile isolates by using multiple-locus variable-number tandem-repeat analysis. J Clin Microbiol 45:1024–8. doi:10.1128/JCM.02023-06
Dridi L, Tankovic J, Burghoffer B, Barbut F, Petit J-C (2002) gyrA and gyrB mutations are implicated in cross-resistance to ciprofloxacin and moxifloxacin in Clostridium difficile. Antimicrob Agents Chemother 46:3418–21. doi:10.1128/aac.46.11.3418-3421.2002
Johnson S, Samore MH, Farrow KA, Killgore GE, Tenover FC, Lyras D, Rood JI, DeGirolami P, Baltch AL, Rafferty ME, Pear SM, Gerding DN (1999) Epidemics of diarrhea caused by a clindamycin-resistant strain of Clostridium difficile in four hospitals. N Engl J Med 341:1645–51. doi:10.1056/NEJM199911253412203
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–9. doi:10.1093/molbev/mst197
Salipante SJ, Hall BG (2011) Inadequacies of minimum spanning trees in molecular epidemiology. J Clin Microbiol 49:3568–75. doi:10.1128/JCM.00919-11
Bastian, M, Heymann, S, Jacomy, M (2009) Gephi: an open source software for exploring and manipulating networks [Internet]. 2009. doi: http://www.aaai.org/ocs/index.php/ICWSM/09/paper/view/154. Accessed 02 July 2016
Eyre DW, Fawley WN, Best EL, Griffiths D, Stoesser NE, Crook DW, Peto TEA, Walker AS, Wilcox MH (2013) Comparison of multilocus variable-number tandem-repeat analysis and whole-genome sequencing for investigation of Clostridium difficile transmission. J Clin Microbiol 51:4141–9. doi:10.1128/JCM.01095-13
Clinical and Laboratory Standards Institute (2014) Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. CLSI document M100-S24, 1st edn. Clinical and Laboratory Standards Institute, Wayne, IN
EUCAST (2016) The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 6.0, 2016 (http://www.eucast.org).
Baines SD, O’Connor R, Freeman J, Fawley WN, Harmanus C, Mastrantonio P, Kuijper EJ, Wilcox MH (2008) Emergence of reduced susceptibility to metronidazole in Clostridium difficile. J Antimicrob Chemother 62:1046–52. doi:10.1093/jac/dkn313
Purcell EB, McKee RW, McBride SM, Waters CM, Tamayo R (2012) Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J Bacteriol 194:3307–16. doi:10.1128/JB.00100-12
Tasteyre A, Barc MC, Karjalainen T, Dodson P, Hyde S, Bourlioux P, Borriello P (2000) A Clostridium difficile gene encoding flagellin. Microbiol Read Engl 146(Pt 4):957–66. doi:10.1099/00221287-146-4-957
Dawson LF, Valiente E, Faulds-Pain A, Donahue EH, Wren BW (2012) Characterisation of Clostridium difficile biofilm formation, a role for Spo0A. PLoS One 7:e50527. doi:10.1371/journal.pone.0050527
Merrigan MM, Venugopal A, Roxas JL, Anwar F, Mallozzi MJ, Roxas BAP, Gerding DN, Viswanathan VK, Vedantam G (2013) Surface-layer protein A (SlpA) is a major contributor to host-cell adherence of Clostridium difficile. PLoS One 8:e78404. doi:10.1371/journal.pone.0078404
Razaq N, Sambol S, Nagaro K, Zukowski W, Cheknis A, Johnson S, Gerding DN (2007) Infection of hamsters with historical and epidemic BI types of Clostridium difficile. J Infect Dis 196:1813–9. doi:10.1086/523106
Sambol SP, Merrigan MM, Lyerly D, Gerding DN, Johnson S (2000) Toxin gene analysis of a variant strain of Clostridium difficile that causes human clinical disease. Infect Immun 68:5480–7. doi:10.1128/iai.68.10.5480-5487.2000
Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, Kuijper EJ, Wilcox MH (2010) The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev 23:529–49. doi:10.1128/CMR.00082-09
Moura I, Spigaglia P, Barbanti F, Mastrantonio P (2013) Analysis of metronidazole susceptibility in different Clostridium difficile PCR ribotypes. J Antimicrob Chemother 68:362–5. doi:10.1093/jac/dks420
National Health Laboratory Service (2013) Western Cape academic hospitals antimicrobial recommendations [Internet]. doi: http://www.fidssa.co.za/Content/Documents/Antibiotic_Rec_2013_final.pdf
WHO (2015) Tuberculosis country profiles [Internet]. WHO. doi: http://www.who.int/tb/country/data/profiles/en/. Accessed 02 July 2016
Wang P, Zhou Y, Wang Z, Xie S, Zhang T, Lin M, Li R, Tan J, Chen Y, Jiang B (2014) Identification of Clostridium difficile ribotype 027 for the first time in Mainland China. Infect Control Hosp Epidemiol 35:95–8. doi:10.1086/674405
Choi YJ, Kim HG, Choi YA, Joo WC, Son DW, Kim CH, Shin YW, Kim YS (2009) A case of pseudomembranous colitis associated with rifampicin therapy in a patient with rectal cancer and gastrointestinal tuberculosis. Korean J Gastroenterol Taehan Sohwagi Hakhoe Chi 53:53–6
Jung S-W, Jeon S-W, Do B-H, Kim S-G, Ha S-S, Cho C-M, Tak W-Y, Kweon Y-O, Kim S-K, Choi Y-H, Cha S-I (2007) Clinical aspects of rifampicin-associated pseudomembranous colitis. J Clin Gastroenterol 41:38–40. doi:10.1097/MCG.0b013e31802dfaf7
Tae CH, Jung S-A, Song HJ, Kim S-E, Choi HJ, Lee M, Hwang Y, Kim H, Lee K (2009) The first case of antibiotic-associated colitis by Clostridium difficile PCR ribotype 027 in Korea. J Korean Med Sci 24:520–4. doi:10.3346/jkms.2009.24.3.520
Sun Y-X, Zhao Y-T, Teng L-L, Ge J-L, Jiang H, Shao L (2013) Clostridium difficile infection associated with antituberculous agents in a patient with tuberculous pericarditis. Int Med Tokyo Jpn 52:1495–7. doi:10.2169/internalmedicine.52.0162
Obuch-Woszczatyński P, Dubiel G, Harmanus C, Kuijper E, Duda U, Wultańska D, van Belkum A, Pituch H (2013) Emergence of Clostridium difficile infection in tuberculosis patients due to a highly rifampicin-resistant PCR ribotype 046 clone in Poland. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol 32:1027–30. doi:10.1007/s10096-013-1845-5
Chang KC, Leung CC, Yew WW, Lam FM, Ho PL, Chau CH, Cheng VCC, Yuen KY (2009) Analyses of fluoroquinolones and Clostridium difficile-associated diarrhoea in tuberculosis patients. Int J Tuberc Lung Dis Off J Int Union Tuberc Lung Dis 13:341–6
Lee YM, Huh KC, Yoon SM, Jang BI, Shin JE, Koo HS, Jung Y, Kim SH, Moon HS, Lee SW, Daejeon-Chungchung Intestinal Research Group (2015) Incidence and clinical outcomes of Clostridium difficile infection after treatment with tuberculosis medication. Gut Liver 10(2):250–254. doi:10.5009/gnl14435
Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, Croucher N, Mastroeni P, Scott P, Raisen C, Mottram L, Fairweather NF, Wren BW, Parkhill J, Dougan G (2009) Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun 77:3661–9. doi:10.1128/IAI.00558-09
Bordeleau E, Purcell EB, Lafontaine DA, Fortier L-C, Tamayo R, Burrus V (2015) Cyclic di-GMP riboswitch-regulated type IV pili contribute to aggregation of Clostridium difficile. J Bacteriol 197:819–32. doi:10.1128/JB.02340-14
Ðapa T, Leuzzi R, Ng YK, Baban ST, Adamo R, Kuehne SA, Scarselli M, Minton NP, Serruto D, Unnikrishnan M (2013) Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J Bacteriol 195:545–55. doi:10.1128/JB.01980-12
Acknowledgments
The authors would like to thank Dr Yolandi Roodt and Lesley Jafta for technical assistance with the capillary gel electrophoresis experiments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by the National Research Foundation of South Africa and the South African Medical Research Council. B. Kullin acknowledges the Claude Leon Foundation and the Carnegie Corporation of New York for Postdoctoral Fellowships and an assistance grant from the European Society of Clinical Microbiology and Infectious Diseases to attend the ANAEROBE 2014 Congress. T. Brock was supported by the Equity Development Programme, UCT.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval and Informed consent
The study was approved by the Ethics Committee of the University of Cape Town (HREC number: 310/2008) and included the obtaining of informed consent of patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Minimum spanning tree representation of MLVA data for C. difficile isolates. Unique MLVA types are represented by individual circles, coloured according to ribotype with the size of the circle representing the number of isolates per MLVA type. Circle labels indicate isolation date with the patient ward in brackets. Numbers between the circles represent the summed tandem repeats (STRDs) between isolates. Isolates with a STRD ≤ 2 are shaded and ribotype 017 strains are enclosed by a dotted line. The tree has been redrawn for ease of viewing and is not to scale (PDF 195 kb)
Rights and permissions
About this article
Cite this article
Kullin, B., Brock, T., Rajabally, N. et al. Characterisation of Clostridium difficile strains isolated from Groote Schuur Hospital, Cape Town, South Africa. Eur J Clin Microbiol Infect Dis 35, 1709–1718 (2016). https://doi.org/10.1007/s10096-016-2717-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-016-2717-6