Abstract
Diphenyl ether derivatives are good candidates for anti-tuberculosis agents that display a promising potency for inhibition of InhA, an essential enoyl-acyl carrier protein (ACP) reductase involved in fatty acid biosynthesis pathways in Mycobacterium tuberculosis. In this work, key structural features for the inhibition were identified by 3D-QSAR CoMSIA models, constructed based on available experimental binding properties of diphenyl ether inhibitors, and a set of four representative compounds was subjected to MD simulations of inhibitor-InhA complexes for the calculation of binding free energies. The results show that bulky groups are required for the R1 substituent on the phenyl A ring of the inhibitors to favor a hydrophobic pocket formed by residues Phe149, Met155, Pro156, Ala157, Tyr158, Pro193, Met199, Val203, Leu207, Ile215, and Leu218. Small substituents with a hydrophilic property are required at the R3 and R4 positions of the inhibitor phenyl B rings to form hydrogen bonds with the backbones of Gly96 and Met98, respectively. For the R2 substituent, small substituents with simultaneous hydrophilic or hydrophobic properties are required to favor the interaction with the pyrophosphate moiety of NAD+ and the methyl side chain of Ala198, respectively. The reported data provide structural guidance for the design of new and potent diphenyl ether-based inhibitors with high inhibitory activities against M. tuberculosis InhA.

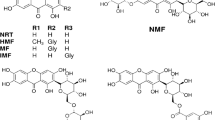
The superimposition of compounds 17 (stick in cyan color), 18 (stick in yellow color), 19 (stick in green color) and 29 (stick in pink color) in the InhA pocket obtained from MD simulation







Similar content being viewed by others
References
World Health Organization (2013) Global tuberculosis report, Available at http://apps.who.int/iris/bitstream/10665/91355/1/9789241564656_eng.pdf
Quemard A, Sacchettini JC, Dessen A, Vilcheze C, Bittman R, Jacobs WR Jr, Blanchard JS (1995) Enzymatic characterization of the target for isoniazid in Mycobacterium tuberculosis. Biochemistry 34(36):8235–8241
Rozwarski DA, Vilchèze C, Sugantino M, Bittman R, Sacchettini JC (1999) Crystal structure of the Mycobacterium tuberculosis enoyl-ACP reductase, InhA, in complex with NAD+ and a C16 fatty acyl substrate. J Biol Chem 274(22):15582–15589
Agüero F, Al-Lazikani B, Aslett M, Berriman M, Buckner FS, Campbell RK, Carmona S, Carruthers IM, Chan AW, Chen F, Crowther GJ, Doyle MA, Hertz-Fowler C, Hopkins AL, McAllister G, Nwaka S, Overington JP, Pain A, Paolini GV, Pieper U, Ralph SA, Riechers A, Roos DS, Sali A, Shanmugam D, Suzuki T, Van Voorhis WC, Verlinde CL (2008) Genomic-scale prioritization of drug targets: the TDR targets database. Nat Rev Drug Discov 7(11):900–907
Campbell JW, Cronan JE Jr (2001) Bacterial fatty acid biosynthesis: targets for antibacterial drug discovery. Annu Rev Microbiol 55:305–332
Heath RJ, Rock CO (2004) Fatty acid biosynthesis as a target for novel antibacterials. Curr Opin Invest Drugs 5(2):146–153
White SW, Zheng J, Zhang YM, Rock CO (2005) The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem 74:791–831
Zhang YM, Lu YJ, Rock CO (2004) The reductase steps of the type II fatty acid synthase as antimicrobial targets. Lipids 39(11):1055–1060
Wen L, Chmielowski JN, Bohn KC, Huang JK, Timsina YN, Kodali P, Pathak AK (2009) Functional expression of Francisella tularensis FabH and FabI, potential antibacterial targets. Protein Exp Purif 65(1):83–91
Wright HT, Reynolds KA (2007) Antibacterial targets in fatty acid biosynthesis. Curr Opin Microbiol 10(5):447–453
Rozwarski DA, Grant GA, Barton DH, Jacobs WR Jr, Sacchettini JC (1998) Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science 279(5347):98–102
Vilcheze C, Wang F, Arai M, Hazbon MH, Colangeli R, Kremer L, Weisbrod TR, Alland D, Sacchettini JC, Jacobs WR Jr (2006) Transfer of a point mutation in Mycobacterium tuberculosis InhA resolves the target of isoniazid. Nat Med 12(9):1027–1029
Dessen A, Quemard A, Blanchard JS, Jacobs WR Jr, Sacchettini JC (1995) Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis. Science 267(5204):1638–1641
Johnsson K, Schultz PG (1994) Mechanistic studies of the oxidation of isoniazid by the catalase peroxidase from Mycobacterium tuberculosis. J Am Chem Soc 116(16):7425–7426
Lei B, Wei CJ, Tu SC (2000) Action mechanism of antitubercular isoniazid. Activation by Mycobacterium tuberculosis KatG, isolation, and characterization of InhA inhibitor. J Biol Chem 275(4):2520–2526
Johnsson K, King DS, Schultz PG (1995) Studies on the mechanism of action of isoniazid and ethionamide in the chemotherapy of tuberculosis. J Am Chem Soc 117(17):5009–5010
Zhang Y, Heym B, Allen B, Young D, Cole S (1992) The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358(6387):591–593
Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, Collins D, de Lisle G, Jacobs WR Jr (1994) InhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263(5144):227–230
Quemard A, Dessen A, Sugantino M, Jacobs WR Jr, Sacchetini JC, Blanchard JS (1996) Binding of catalase-peroxidase-activated isoniazid to wild-type and mutant Mycobacterium tuberculosis enoyl-ACP reductases. J Am Chem Soc 118(6):1561–1562
Saint-Joanis B, Souchon H, Wilming M, Johnsson K, Alzari PM, Cole ST (1999) Use of site-directed mutagenesis to probe the structure, function and isoniazid activation of the catalase/peroxidase, KatG, from Mycobacterium tuberculosis. Biochem J 338(Pt 3):753–760
Zhao X, Yu H, Yu S, Wang F, Sacchettini JC, Magliozzo RS (2006) Hydrogen peroxide-mediated isoniazid activation catalyzed by Mycobacterium tuberculosis catalase-peroxidase (KatG) and its S315T mutant. Biochemistry 45(13):4131–4140
Metcalfe C, Macdonald IK, Murphy EJ, Brown KA, Raven EL, Moody PC (2008) The tuberculosis prodrug isoniazid bound to activating peroxidases. J Biol Chem 283(10):6193–6200
Sinha BK (1983) Enzymatic activation of hydrazine derivatives. J Biol Chem 258(2):796–801
Nguyen M, Claparols C, Bernadou J, Meunier B (2001) A fast and efficient metal-mediated oxidation of isoniazid and identification of isoniazid-NAD(H) adducts. ChemBioChem 2(12):877–883
Heym B, Zhang Y, Poulet S, Young D, Cole ST (1993) Characterization of the katG gene encoding a catalase-peroxidase required for the isoniazid susceptibility of Mycobacterium tuberculosis. J Bacteriol 175(13):4255–4259
Timmins GS, Deretic V (2006) Mechanisms of action of isoniazid. Mol Microbiol 62(5):1220–1227
Johnsson K, Froland WA, Schultz PG (1997) Overexpression, purification, and characterization of the catalase-peroxidase KatG from Mycobacterium tuberculosis. J Biol Chem 272(5):2834–2840
De La Iglesia AI, Morbidoni HR (2006) Mechanisms of action of and resistance to rifampicin and isoniazid in Mycobacterium tuberculosis: new information on old friends. Rev Argent Microbiol 38(2):97–109
Parikh SL, Xiao G, Tonge PJ (2000) Inhibition of InhA, the enoyl reductase from Mycobacterium tuberculosis, by triclosan and isoniazid. Biochemistry 39(26):7645–7650
Freundlich JS, Wang F, Vilchèze C, Gulten G, Langley R, Schiehser GA, Jacobus DP, Jacobs WR Jr, Sacchettini JC (2009) Triclosan derivatives: towards potent inhibitors of drug-sensitive and drug-resistant Mycobacterium tuberculosis. ChemMedChem 4(2):241–248
am Ende CW, Knudson SE, Liu N, Childs J, Sullivan TJ, Boyne M, Xu H, Gegina Y, Knudson DL, Johnson F, Peloquin CA, Slayden RA, Tonge PJ, Ende CW, Knudson SE, Liu N, Childs J, Sullivan TJ, Boyne M, Xu H, Gegina Y, Knudson DL, Johnson F, Peloquin CA, Slayden RA, Tonge PJ (2008) Synthesis and in vitro antimycobacterial activity of B-ring modified diaryl ether InhA inhibitors. Bioorg Med Chem Lett 18(10):3029–3033
Boyne ME, Sullivan TJ, am Ende CW, Lu H, Gruppo V, Heaslip D, Amin AG, Chatterjee D, Lenaerts A, Tonge PJ, Slayden RA (2007) Targeting fatty acid biosynthesis for the development of novel chemotherapeutics against Mycobacterium tuberculosis: evaluation of A-ring-modified diphenyl ethers as high-affinity InhA inhibitors. Antimicrob Agents Chemother 51(10):3562–3567
Sullivan TJ, Truglio JJ, Boyne ME, Novichenok P, Zhang X, Stratton CF, Li HJ, Kaur T, Amin A, Johnson F, Slayden RA, Kisker C, Tonge PJ (2006) High affinity InhA inhibitors with activity against drug-resistant strains of Mycobacterium tuberculosis. ACS Chem Biol 1(1):43–53
Luckner SR, Liu N, am Ende CW, Tonge PJ, Kisker C (2010) A slow, tight-binding inhibitor of InhA, the enoyl-ACP reductase from Mycobacterium tuberculosis. J Biol Chem 285(19):14330–14337
Pan P (2012) Lead optimization and slow-onset inhibition of the enoyl-ACP reductase (InhA) from Mycobacterium tuberculosis. PhD Diss., Stony Brook University
Lu XY, Chen YD, Jiang YJ, You QD (2009) Discovery of potential new InhA direct inhibitors based on pharmacophore and 3D-QSAR analysis followed by in silico screening. Eur J Med Chem 44(9):3718–3730
Kumar A, Siddiqi MI (2008) CoMFA based de novo design of pyrrolidine carboxamides as inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis. J Mol Model 14(10):923–935
Andrade CH, Salum Lde B, Castilho MS, Pasqualoto KF, Ferreira EI, Andricopulo AD (2008) Fragment-based and classical quantitative structure-activity relationships for a series of hydrazides as antituberculosis agents. Mol Divers 12(1):47–59
Lu XU, Chen YD, You QD (2010) 3D-QSAR studies of arylcarboxamides with inhibitory activity on InhA using pharmacophore-based alignment. Chem Bio Drug Des 75(2):195–203
Punkvang A, Saparpakorn P, Hannongbua S, Wolschann P, Beyer A, Pungpo P (2010) Investigating the structural basis of arylamides to improve potency against M. tuberculosis strain through molecular dynamics simulations. Eur J Med Chem 45(12):5585–5593
Punkvang A, Saparpakorn P, Hannongbua S, Wolschann P, Pungpo P (2010) Elucidating drug-enzyme interactions and their structural basis for improving the affinity and potency of isoniazid and its derivatives based on computer modeling approaches. Molecules 15(4):2791–2813
Punkvang A, Saparpakorn P, Hannongbua S, Wolschann P, Berner H, Pungpo P (2010) Insight into crucial inhibitor–enzyme interaction of arylamides as novel direct inhibitors of the enoyl ACP reductase (InhA) from Mycobacterium tuberculosis: computer-aided molecular design. Monatsh Chem 141(9):1029–1041
Pauli I, dos Santos RN, Rostirolla DC, Martinelli LK, Ducati RG, Timmers LF, Basso LA, Santos DS, Guido RV, Andricopulo AD, Norberto de Souza O (2013) Discovery of new inhibitors of Mycobacterium tuberculosis InhA enzyme using virtual screening and a 3D-pharmacophore-based approach. J Chem Inf Model 53(9):2390–2401
Kinjo T, Koseki Y, Kobayashi M, Yamada A, Morita K, Yamaguchi K, Tsurusawa R, Gulten G, Komatsu H, Sakamoto H, Sacchettini JC, Kitamura M, Aoki S (2013) Identification of compounds with potential antibacterial activity against Mycobacterium through structure-based drug screening. J Chem Inf Model 53(5):1200–1212
Kumar UC, Bvs SK, Mahmood S, Sriram D, Kumar-Sahu P, Pulakanam S, Ballell L, Alvarez-Gomez D, Malik S, Jarp S (2013) Discovery of novel InhA reductase inhibitors: application of pharmacophore- and shape-based screening approach. Future Med Chem 5(3):249–259
Stigliani JL, Bernardes-Génisson V, Bernadou J, Pratviel G (2012) Cross-docking study on InhA inhibitors: a combination of Autodock Vina and PM6-DH2 simulations to retrieve bio-active conformations. Org Biomol Chem 10:6341–6349
Subba Rao G, Vijayakrishnan R, Kumar M (2008) Structure-based design of a novel class of potent inhibitors of InhA, the enoyl acyl carrier protein reductase from Mycobacterium tuberculosis: a computer modelling approach. Chem Biol Drug Des 72(5):444–449
Stigliani JL, Arnaud P, Delaine T, Bernardes-Génisson V, Meunier B, Bernadou J (2008) Binding of the tautomeric forms of isoniazid-NAD adducts to the active site of the Mycobacterium tuberculosis enoyl-ACP reductase (InhA): a theoretical approach. J Mol Graph Model 27(4):536–545
(2006) GaussView 03, Revision 3.07, Gaussian, Inc., Wallingford, CT
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Jr. JAM, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas O, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ (2009) Gaussian 09, revision B.01, Gaussian Inc., Wallingford CT
Morris GM, Goodsell DS, Huey R, Olson AJ (1996) Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J Comput Aided Mol Des 10(4):293–304
Tripos International (2008) SYBYL v8.0, Tripos International, 1699 South Hanley Rd., St. Louis, MO
Golbraikh A, Tropsha A (2002) Beware of q2! J Mol Graph Model 20:267–276
Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA (2008) AMBER12. University of California, San Francisco
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24(16):1999–2012
Wang J, Wang W, Kollman PA, Case DA (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graphics Model 25(2):247–260
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general AMBER force field. J Comput Chem 25(9):1157–1174
Cornell WD, Cieplak P, Bayly CI, Kollman PA (1993) Application of RESP charges to calculation conformational energies, hydrogen bond energies, and free energies of salvation. J Am Chem Soc 115(21):9620–9631
Bayly CI, Cieplak P, Cornell WD, Kollman PA (1993) A well-behaved electrostatic potential based method using charge-restraints for deriving charges: The RESP model. J Phys Chem 97(40):10269–10280
Wang J, Cieplak P, Kollman PA (2000) How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem 21(12):1049–1074
Gavernet L, Gonzalez Funes JL, Blanch LB, Estiu G, Maresca A, Supuran CT (2010) Affinity of sulfamates and sulfamides to carbonic anhydrase II isoform: experimental and molecular modeling approaches. J Chem Inf Model 50(6):1113–1122
Li W, Fu J, Cheng F, Zheng M, Zhang J, Liu G, Tang Y (2012) Unbinding pathways of GW4064 from human farnesoid X receptor as revealed by molecular dynamics simulations. J Chem Inf Model 52(11):3043–3052
Mahoney MW, Jorgensen WL (2000) A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J Chem Phys 112(20):8910–8922
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J Chem Phys 98(12):10089–10092
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comp Phys 23(3):327–341
Homeyer N, Gohlke H (2012) Free energy calculations by the molecular mechanics Poisson–Boltzmann surface area method. Mol Inf 31(2):114–122
Wang J, Hou T, Xu X (2006) Recent advances in free energy calculations with a combination of molecular mechanics and continuum models. Curr Comput Aided Drug Des 2(3):95–103
Wang J, Morin P, Wang W, Kollman PA (2001) Use of MM-PBSA in reproducing the binding free energies to HIV1 RT of TIBO derivatives and predicting the binding mode to HIV1 RT of Efavirenz by docking and MM-PBSA. J Am Chem Soc 123(22):5221–5230
Hou T, Wang J, Li Y, Wang W (2011) Assessing the performance of the MM/PBSA and MM/GBSA Methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 51(1):69–82
Kaledin M, Brown A, Kaledin AL, Bowman JM (2004) Normal mode analysis using the driven molecular dynamics method. II. An application to biological macromolecules. J Chem Phys 121(12):5646–5653
Xue W, Qi J, Yang Y, Jin X, Liu H, Yao X (2012) Understanding the effect of drug-resistant mutations of HIV-1 intasome on raltegravir action through molecular modeling study. Mol BioSyst 8:2135–2144
Zar JH (1998) Biostatistical analysis. Prentice Hall, Upper Saddle River
Acknowledgments
This research has been supported by the Thailand Research Fund (DBG5380006, MRG5680169, and RTA5380010), the National Research Council of Thailand and Higher Education Research Promotion. The financial support from Royal Golden Jubilee Ph.D. Program (PHD/004/2554) to P. Kamsri is gratefully acknowledged. Faculty of Science, Ubon Ratchathani University, University of Vienna, ASEA-Uninet (Austrian - South-East Asian University Partnership Network), NECTEC (National Electronics and Computer Technology Center) and BIOTEC (National center for genetic engineering and biotechnology) are gratefully acknowledged for supporting this research.
Author information
Authors and Affiliations
Corresponding author
Additional information
This paper belongs to Topical Collection 9th European Conference on Computational Chemistry (EuCo-CC9)
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 695 kb)
Rights and permissions
About this article
Cite this article
Kamsri, P., Punkvang, A., Saparpakorn, P. et al. Elucidating the structural basis of diphenyl ether derivatives as highly potent enoyl-ACP reductase inhibitors through molecular dynamics simulations and 3D-QSAR study. J Mol Model 20, 2319 (2014). https://doi.org/10.1007/s00894-014-2319-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-014-2319-0