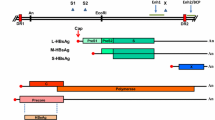
Abstract
The most characteristic feature of the hepatitis C virus (HCV) genome in patients with chronic hepatitis C is its remarkable variability and diversity. To better understand this feature, we performed genetic analysis of HCV replicons recovered from two human hepatoma HuH-7-derived cell lines after 1, 3, 5, 7, and 9 years in culture: The cell lines 50-1 and sO harbored HCV 1B-1 and O strain-derived HCV replicons established in 2002 and 2003, respectively. The results revealed that genetic variations in both replicons accumulated in a time-dependent manner at a constant rate despite the maintenance of moderate diversity (less than 1.8% difference) between the clones and that the mutation rate in the 50-1 and sO replicons was 2.5 and 2.9 × 10−3 base substitutions/site/year, respectively. We found that the genetic distance of both replicons increased from 7.9% to 10.5% after 9 years in culture. In addition, we observed that the guanine + cytosine (GC) content of both replicon RNAs increased in a time-dependent manner, as observed in our previous studies. Finally, we demonstrated that the high sensitivity of both replicons to direct-acting antivirals was maintained even after 9 years in culture. Our results suggest that long-term cultured HCV replicon-harboring cells are a useful model for understanding the variability and diversity of the HCV genome and the drug sensitivity of HCV in patients with chronic hepatitis C.






Similar content being viewed by others
References
Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K (1990) Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA 87:9524–9528
Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y, Choo QL, Houghton M, Kuo G (1990) Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA 87:6547–6549
Kato N (2001) Molecular virology of hepatitis C virus. Acta Med Okayama 55:133–159
Dubuisson J, Cosset FL (2014) Virology and cell biology of the hepatitis C virus life cycle: an update. J Hepatol 61:S3–S13
Suzuki T (2017) Hepatitis C virus replication. Adv Exp Med Biol 997:199–209
Pawlotsky JM (2016) Hepatitis C virus resistance to direct-acting antiviral drugs in interferon-free regimens. Gastroenterology 151:70–86
Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R (1999) Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113
Ikeda M, Yi M, Li K, Lemon SM (2002) Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J Virol 76:2997–3006
Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M, Bartenschlager R, Liang TJ (2005) Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796
Kishine H, Sugiyama K, Hijikata M, Kato N, Takahashi H, Noshi T, Nio Y, Hosaka M, Miyanari Y, Shimotohno K (2002) Subgenomic replicon derived from a cell line infected with the hepatitis C virus. Biochem Biophys Res Commun 293:993–999
Kato N, Sugiyama K, Namba K, Dansako H, Nakamura T, Takami M, Naka K, Nozaki A, Shimotohno K (2003) Establishment of a hepatitis C virus subgenomic replicon derived from human hepatocytes infected in vitro. Biochem Biophys Res Commun 306:756–766
Ogata N, Alter HJ, Miller RH, Purcell RH (1991) Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci USA 88:3392–3396
Okamoto H, Kojima M, Okada S, Yoshizawa H, Iizuka H, Tanaka T, Muchmore EE, Peterson DA, Ito Y, Mishiro S (1992) Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology 190:894–899
Major ME, Mihalik K, Fernandez J, Seidman J, Kleiner D, Kolykhalov AA, Rice CM, Feinstone SM (1999) Long-term follow-up of chimpanzees inoculated with the first infectious clone for hepatitis C virus. J Virol 73:3317–3325
Tanaka T, Kato N, Nakagawa M, Ootsuyama Y, Cho MJ, Nakazawa T, Hijikata M, Ishimura Y, Shimotohno K (1992) Molecular cloning of hepatitis C virus genome from a single Japanese carrier: sequence variation within the same individual and among infected individuals. Virus Res 23:39–53
Kato N, Nakamura T, Dansako H, Namba K, Abe K, Nozaki A, Naka K, Ikeda M, Shimotohno K (2005) Genetic variation and dynamics of hepatitis C virus replicons in long-term cell culture. J Gen Virol 86:645–656
Ikeda M, Abe K, Dansako H, Nakamura T, Naka K, Kato N (2005) Efficient replication of a full-length hepatitis C virus genome, strain O, in cell culture, and development of a luciferase reporter system. Biochem Biophys Res Commun 329:1350–1359
Abe K, Ikeda M, Dansako H, Naka K, Kato N (2007) Cell culture-adaptive NS3 mutations required for the robust replication of genome-length hepatitis C virus RNA. Virus Res 125:88–97
Kato N, Abe K, Mori K, Ariumi Y, Dansako H, Ikeda M (2009) Genetic variability and diversity of intracellular genome-length hepatitis C virus RNA in long-term cell culture. Arch Virol 154:77–85
Kato N, Sejima H, Ueda Y, Mori K, Satoh S, Dansako H, Ikeda M (2014) Genetic characterization of hepatitis C virus in long-term RNA replication using Li23 cell culture systems. PLoS One 9:e91156
Kato N, Mori K, Abe K, Dansako H, Kuroki M, Ariumi Y, Wakita T, Ikeda M (2009) Efficient replication systems for hepatitis C virus using a new human hepatoma cell line. Virus Res 146:41–50
Pietschmann T, Lohmann V, Kaul A, Krieger N, Rinck G, Rutter G, Strand D, Bartenschlager R (2002) Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J Virol 76:4008–4021
Ueda Y, Dansako H, Satoh S, Kim HS, Wataya Y, Doi H, Ikeda M, Kato N (2017) Evaluation of preclinical antimalarial drugs, which can overcome direct-acting antivirals-resistant hepatitis C viruses, using the viral reporter assay systems. Virus Res 235:37–48
Moreno E, Gallego I, Gregori J, Lucía-Sanz A, Soria ME, Castro V, Beach NM, Manrubia S, Quer J, Esteban JI, Rice CM, Gómez J, Gastaminza P, Domingo E, Perales C (2017) Internal disequilibria and phenotypic diversification during replication of hepatitis C virus in a noncoevolving cellular environment. J Virol 91:e02505–e02516
Weiner A, Erickson AL, Kansopon J, Crawford K, Muchmore E, Hughes AL, Houghton M, Walker CM (1995) Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc Natl Acad Sci USA 92:2755–2759
Tan SL, editor (2006) Hepatitis C viruses: Genomes and Molecular Biology. Norfolk (UK): Horizon Bioscience. http://www.ncbi.nlm.nih.gov/books/NBK1613/. Accessed 10 Feb 2014
Dietz J, Susser S, Vermehren J, Peiffer KH, Grammatikos G, Berger A, Ferenci P, Buti M, Müllhaupt B, Hunyady B, Hinrichsen H, Mauss S, Petersen J, Buggisch P, Felten G, Hüppe D, Knecht G, Lutz T, Schott E, Berg C, Spengler U, von Hahn T, Berg T, Zeuzem S, Sarrazin C, European HCV Resistance Study Group (2018) Patterns of resistance-associated substitutions in patients with chronic HCV infection following treatment with direct-acting antivirals. Gastroenterology 154:976–988
Acknowledgements
We thank Takashi Nakamura, Keiko Takeshita, Narumi Yamane, and Naoko Kimura for their technical assistance. We also thank Dr. Kyoko Mori, Dr. Yasuo Ariumi, Dr. Ken-ichi Abe, Dr. Kazuhito Naka, Dr. Akito Nozaki, and Dr. Kazuo Sugiyama for their helpful suggestions. This study was supported by a JSPS KAKENHI grant (no. 16H05196) and a grant for Practical Research on Hepatitis from the Japan Agency for Medical Research and Development (AMED) (no. JP19fk0210053h0301).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Handling Editor: Michael A. Purdy.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Genetic variations occurring during the 9-year replication of HCV 50-1 and sO replicon RNAs. (A) Genetic variations in the first 2-kb region of the HCV 50-1 replicon. The horizontal line indicates the culture time (in years). The genetic distance (%) of the clones obtained from 50-1(1Y), 50-1(3Y), 50-1(5Y), 50-1(7Y), and 50-1(9Y) cells from the 50-1(0Y) cell-derived HCV replicon [16] is shown in the figure. (B) Genetic variations in the first 2-kb region of the HCV sO replicon. The genetic distance (%) of the clones obtained from sO(1Y), sO(3Y), sO(5Y), sO(7Y), and sO(9Y) cells from the sO(0Y) cell-derived HCV replicon [11] is shown in the figure (TIFF 1241 kb)
Fig. S2
Phylogenetic trees based on deduced amino acid sequences of the NS 6-kb regions of HCV 50-1 and sO replicon populations obtained over a 9-year time period. (A) Phylogenetic tree of the NS 6-kb region of the HCV 50-1 replicon based on the deduced 1987-amino-acid sequences of all cDNA clones obtained from 50-1(1Y), 50-1(3Y), 50-1(5Y), 50-1(7Y), and 50-1(9Y) cells. 50-1(0Y, Origin) indicates the original sequences of the NS 6-kb region of the HCV 50-1 replicon [10]. (B) Phylogenetic tree of the NS 6-kb region of the HCV sO replicon based on the deduced 1987-amino-acid sequences of all cDNA clones obtained from sO(1Y), sO(3Y), sO(5Y), sO(7Y), and sO(9Y) cells. sO(0Y, Origin) indicates the original sequences of the NS 6 kb region of the HCV sO replicon [11] (TIFF 1350 kb)
705_2019_4461_MOESM3_ESM.tif
Phylogenetic trees of HCV 50-1 and sO replicon populations obtained over a 9-year period. (A) Phylogenetic tree of the first 2-kb region of the HCV 50-1 replicon based on the nucleotide sequences of all cDNA clones obtained from 50-1(1Y), 50-1(3Y), 50-1(5Y), 50-1(7Y), and 50-1(9Y) cells. 50-1(0Y, Origin) indicates the original sequences of the first 2-kb region of the HCV 50-1 replicon [10]. (B) Phylogenetic tree of the first 2-kb region of the HCV sO replicon based on the nucleotide sequences of all cDNA clones obtained from sO(1Y), sO(3Y), sO(5Y), sO(7Y), and sO(9Y) cells. sO(0Y, Origin) indicates the original sequences of the first 2-kb region of the HCV sO replicon [11] (TIFF 1354 kb)
Rights and permissions
About this article
Cite this article
Kato, N., Ueda, Y., Sejima, H. et al. Study of multiple genetic variations caused by persistent hepatitis C virus replication in long-term cell culture. Arch Virol 165, 331–343 (2020). https://doi.org/10.1007/s00705-019-04461-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04461-0