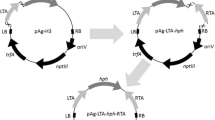
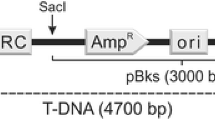
Abstract
Agrocybe aegerita is a cultivated edible mushroom in numerous countries, which also serves as a model basidiomycete to study fruiting body formation. Aiming to create an easily expandable customised molecular toolset for transformation and constitutive gene of interest expression, we first created a homologous dominant marker for transformant selection. Progeny monokaryons of the genome-sequenced dikaryon A. aegerita AAE-3 used here were identified as sensitive to the systemic fungicide carboxin. We cloned the wild-type gene encoding the iron-sulphur protein subunit of succinate dehydrogenase AaeSdi1 including its up- and downstream regions, and introduced a single-point mutation (His237 to Leu) to make it confer carboxin resistance. PEG-mediated transformation of protoplasts derived from either oidia or vegetative monokaryotic mycelium with the resulting carboxin resistance marker (CbxR) plasmid pSDI1E3 yielded carboxin-resistant transformants in both cases. Plasmid DNA linearised within the selection marker resulted in transformants with ectopic multiple insertions of plasmid DNA in a head-to-tail repeat-like fashion. When circular plasmid was used, ectopic single integration into the fungal genome was favoured, but also gene conversion at the homologous locus was seen in 1 out of 11 analysed transformants. Employing CbxR as selection marker, two versions of a reporter gene construct were assembled via Golden Gate cloning which allows easy recombination of its modules. These consisted of an eGFP expression cassette controlled by the native promoter PAaeGPDII and the heterologous terminator Tnos, once with and once without an intron in front of the eGFP start codon. After protoplast transformation with either construct as circular plasmid DNA, GFP fluorescence was detected with either transformants, indicating that expression of eGFP is intron-independent in A. aegerita. This paves the way for functional genetics approaches to A. aegerita, e.g., via constitutive expression of fruiting-related genes.




Similar content being viewed by others
Data availability
The AaeSDI1 gene sequence employed in this study is available from the NCBI (National Center for Biotechnology Information) GenBank® repository via accession number KU984434. DNA sequence information of AaeSDI1 was obtained via web-based genome browser-mediated access to the in silico-annotated genome sequence of A. aegerita AAE-3 (http://www.thines-lab.senckenberg.de/agrocybe_genome), and the parental dikaryon of the monokaryons A. aegerita AAE-3-13 and A. aegerita AAE-3-32 (Herzog et al. 2016) which were used throughout this study. The genome sequence data of A. aegerita AAE-3 have also been deposited in the European Nucleotide Archive (ENA) under the BioProject accession number PRJEB21917.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Amore A, Honda Y, Faraco V (2012) Enhanced green fluorescent protein expression in Pleurotus ostreatus for in vivo analysis of fungal laccase promoters. Appl Biochem Biotechnol 168:761–769
Böhmer C, Böhmer M, Bölker M, Sandrock B (2008) Cdc42 and the Ste20-like kinase Don3 act independently in triggering cytokinesis in Ustilago maydis. J Cell Sci 121:143–148
Brachmann A (2001) Die frühe Infektionsphase von Ustilago maydis: genregulation durch das bW/bE-Heterodimer. PhD Thesis, Fakultät für Biologie, LMU München, Munich, Germany
Broomfield PL, Hargreaves JA (1992) A single amino-acid change in the iron-sulphur protein subunit of succinate dehydrogenase confers resistance to carboxin in Ustilago maydis. Curr Genet 22:117–121
Burns C, Gregory KE, Kirby M et al (2005) Efficient GFP expression in the mushrooms Agaricus bisporus and Coprinus cinereus requires introns. Fungal Genet Biol 42:191–199
Burns C, Leach KM, Elliott TJ, Challen MP, Foster GD, Bailey A (2006) Evaluation of agrobacterium-mediated transformation of Agaricus bisporus using a range of promoters linked to hygromycin resistance. Mol Biotechnol 32:129–138
Collins CM, Heneghan MN, Kilaru S, Bailey AM, Foster GD (2010) Improvement of the Coprinopsis cinerea molecular toolkit using new construct design and additional marker genes. J Microbiol Methods 82:156–162
de Jong JF, Deelstra HJ, Wösten HAB, Lugones LG (2006) RNA-mediated gene silencing in monokaryons and dikaryons of Schizophyllum commune. Appl Environ Microbiol 72:1267–1269
de Jong JF, Ohm RA, de Bekker C, Wösten HAB, Lugones LG (2010) Inactivation of ku80 in the mushroom-forming fungus Schizophyllum commune increases the relative incidence of homologous recombination. FEMS Microbiol Lett 310:91–95
Dons JJM, de Vries OMH, Wessels JGH (1979) Characterization of the genome of the basidiomycete Schizophyllum commune. Biochim Biophys Acta 563:100–112
Dörnte B, Kües U (2012) Reliability in transformation of the basidiomycete Coprinopsis cinerea. Curr Trends Biotechnol Pharm 6:340–355
Engler C, Kandzia R, Marillonnet S (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS One 3:e3647
Esser K, Meinhardt F (1977) A common genetic control of dikaryotic and monokaryotic fruiting in the basidiomycete Agrocybe aegerita. Mol Gen Genet 155:113–115
Fincham JRS (1989) Transformation in fungi. Microbiol Rev 53:148–170
Ford KL, Baumgartner K, Henricot B, Bailey AM, Foster GD (2016) A native promoter and inclusion of an intron is necessary for efficient expression of GFP or mRFP in Armillaria mellea. Sci Rep 6:29226
Fotheringham S, Holloman WK (1990) Pathways of transformation in Ustilago Maydis determined by DNA conformation. Genetics 124:833–843
Garcia K, Haider MZ, Delteil A et al (2013) Promoter-dependent expression of the fungal transporter HcPT1.1 under Pi shortage and its spatial localization in ectomycorrhiza. Fungal Genet Biol 58–59:53–61
Godio RP, Fouces R, Gudina EJ, Martin JF (2004) Agrobacterium tumefaciens-mediated transformation of the antitumor clavaric acid-producing basidiomycete Hypholoma sublateritium. Curr Genet 46:287–294
Gupta DK, Rühl M, Mishra B et al (2018) The genome sequence of the commercially cultivated mushroom Agrocybe aegerita reveals a conserved repertoire of fruiting-related genes and a versatile suite of biopolymer-degrading enzymes. BMC Genom 19:48
Gutierrez-Triana JA, Tavhelidse T, Thumberger T et al (2018) Efficient single-copy HDR by 5′ modified long dsDNA donors. eLife 7:e39468
Harmsen MC, Schuren FHJ, Moukha SM, Vanzuilen CM, Punt PJ, Wessels JGH (1992) Sequence-analysis of the glyceraldehyde-3-phosphate dehydrogenase genes from the basidiomycetes Schizophyllum commune, Phanerochaete chrysosporium and Agaricus bisporus. Curr Genet 22:447–454
Herzog R, Solovyeva I, Rühl M, Thines M, Hennicke F (2016) Dikaryotic fruiting body development in a single dikaryon of Agrocybe aegerita and the spectrum of monokaryotic fruiting types in its monokaryotic progeny. Mycol Progress 15:947–957
Honda Y, Matsuyama T, Irie T, Watanabe T, Kuwahara M (2000) Carboxin resistance transformation of the homobasidiomycete fungus Pleurotus ostreatus. Curr Genet 37:209–212
Irie T, Sato T, Saito K et al (2003) Construction of a homologous selectable marker gene for Lentinula edodes transformation. Biosci Biotechnol Biochem 67:2006–2009
Ito Y, Muraguchi H, Seshime Y, Oita S, Yanagi SO (2004) Flutolanil and carboxin resistance in Coprinus cinereus conferred by a mutation in the cytochrome b560 subunit of succinate dehydrogenase complex (Complex II). Mol Genet Genom 272:328–335
Kearse M, Moir R, Wilson A et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Keon JP, White GA, Hargreaves JA (1991) Isolation, characterization and sequence of a gene conferring resistance to the systemic fungicide carboxin from the maize smut pathogen, Ustilago maydis. Curr Genet 19:475–481
Kilaru S, Collins CM, Hartley JH, Burns C, Foster GD, Bailey AM (2009) Investigating dominant selection markers for Coprinopsis cinerea: a carboxin resistance system and re-evaluation of hygromycin and phleomycin resistance vectors. Curr Genet 55:543–550
Kleofas V, Sommer L, Fraatz MA, Zorn H, Rühl M (2014) Fruiting body production and aroma profile analysis of Agrocybe aegerita cultivated on different substrates. Nat Resour 5:233–240
Kües U (2000) Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol Mol Biol Rev 64:316–353
Labarère J, Noël T (1992) Mating type switching in the tetrapolar basidiomycete Agrocybe aegerita. Genetics 131:307–319
Lorang JM, Tuori RP, Martinez JP et al (2001) Green fluorescent protein is lighting up fungal biology. Appl Environ Microbiol 67:1987–1994
Lugones LG, Scholtmeijer K, Klootwijk R, Wessels JGH (1999) Introns are necessary for mRNA accumulation in Schizophyllum commune. Mol Microbiol 32:681–689
Ma B, Mayfield MB, Gold MH (2001) The green fluorescent protein gene functions as a reporter of gene expression in Phanerochaete chrysosporium. Appl Environ Microbiol 67:948–955
Meinhardt F, Esser K (1981) Genetic studies of the basidiomycete Agrocybe aegerita. 2. Genetic control of fruit body formation and its practical implications. Theor Appl Genet 60:265–268
Meinhardt F, Leslie F (1982) Mating Types of Agrocybe aegerita. Curr Genet 5:65–68
Mooibroek H, Kuipers AG, Sietsma JH, Punt PJ, Wessels JG (1990) Introduction of hygromycin B resistance into Schizophyllum commune: preferential methylation of donor DNA. Mol Gen Genet 222:41–48
Müller T, Benjdia M, Avolio M et al (2006) Functional expression of the green fluorescent protein in the ectomycorrhizal model fungus Hebeloma cylindrosporum. Mycorrhiza 16:437–442
Noël T, Labarère J (1992) Sequence of the URA1 gene encoding dihydroorotate dehydrogenase from the basidiomycete fungus Agrocybe aegerita. Gene 122:233–234
Noël T, Labarère J (1994) Homologous transformation of the edible basidiomycete Agrocybe aegerita with the URA1 gene: characterization of integrative events and of rearranged free plasmids in transformants. Curr Genet 25:432–437
Noël T, Simoneau P, Labarère J (1995) Heterologous transformation of Agrocybe aegerita with a bacterial neomycin-resistance gene fused to a fungal promoter-like DNA sequence. Theor Appl Genet 90:1019–1027
Ohm RA, de Jong JF, Lugones LG et al (2010) Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol 28:957–963
Okuda Y, Murakami S, Honda Y, Matsumoto T (2013) An MSH4 homolog. stpp1, from Pleurotus pulmonarius Is a “silver bullet” for resolving problems caused by spores in cultivated mushrooms. Appl Environ Microbiol 79:4520–4527
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sato M, Kurahashi A, Nishibori K, Fujimori F (2014) Development of a transformation system for the edible mushroom Grifola frondosa: demonstrating heterologous gene expression and RNAi-mediated gene silencing. Mycoscience 56:364–372
Scholtmeijer K, Wösten HAB, Springer J, Wessels JG (2001) Effect of introns and AT-rich sequences on expression of the bacterial hygromycin B resistance gene in the basidiomycete Schizophyllum commune. Appl Environ Microbiol 67:481–483
Shi L, Fang X, Li M et al (2012) Development of a simple and efficient transformation system for the basidiomycetous medicinal fungus Ganoderma lucidum. World J Microbiol Biotechnol 28:283–291
Sirand-Pugnet P, Santos C, Labarère J (2003) The Aa-Pri4 gene, specifically expressed during fruiting initiation in the Agrocybe aegerita complex, contains an unusual CT-rich leader intron within the 5′ uncoding region. Curr Genet 44:124–131
Sugano SS, Suzuki H, Shimokita E et al (2017) Genome editing in the mushroom-forming basidiomycete Coprinopsis cinerea, optimized by a high-throughput transformation system. Sci Rep 7:1260
Terfrüchte M, Joehnk B, Fajardo-Somera R et al (2014) Establishing a versatile Golden Gate cloning system for genetic engineering in fungi. Fungal Genet Biol 62:1–10
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Uhart M, Piscera JM, Albertó E (2008) Utilization of new naturally occurring strains and supplementation to improve the biological efficiency of the edible mushroom Agrocybe cylindracea. J Ind Microbiol Biotechnol 35:595–602
van Peer AF, de Bekker C, Vinck A, Wösten HA, Lugones LG (2009) Phleomycin increases transformation efficiency and promotes single integrations in Schizophyllum commune. Appl Environ Microbiol 75:1243–1247
Wang M, Gu B, Huang J et al (2013) Transcriptome and proteome exploration to provide a resource for the study of Agrocybe aegerita. PLoS One 8:e56686
Acknowledgements
This work was funded by the Senckenberg Gesellschaft für Naturforschung and the Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz (LOEWE) by the State of Hesse’s Ministry of Higher Education, Research and the Arts. Dr. Michael Bölker was supported by the DFG-funded SFB 987 ‘Microbial Diversity in Environmental Signal Response’. DNA sequence data were obtained from the genome sequence of Agrocybe aegerita AAE-3 (Gupta et al. 2018; http://www.thines-lab.senckenberg.de/agrocybe_genome), the parental dikaryon of the monokaryons A. aegerita AAE-3-13 and A. aegerita AAE-3-32, which were used throughout this study. We thank Dr. Björn Sandrock (Philipps-University Marburg, Germany) for fruitful exchange of ideas on the modes of plasmid-chromosomal integration of pSDI1E3 (carboxin resistance cassette-containing plasmid created in this study) in A. aegerita.
Funding
This study was funded by the Senckenberg Gesellschaft für Naturforschung and the Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz (LOEWE) by the State of Hesse’s Ministry of Higher Education, Research and the Arts. Dr. Michael Bölker was supported by the DFG-funded SFB 987 ‘Microbial Diversity in Environmental Signal Response’.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: RH, IS, and FH. Performed the experiments: RH, IS, and FH. Analysed the data: RH, IS, LGL, and FH. Wrote the paper: RH, IS, MB, LGL, and FH.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Not applicable.
Additional information
Communicated by Reinhard Fischer.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Herzog, R., Solovyeva, I., Bölker, M. et al. Exploring molecular tools for transformation and gene expression in the cultivated edible mushroom Agrocybe aegerita. Mol Genet Genomics 294, 663–677 (2019). https://doi.org/10.1007/s00438-018-01528-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-018-01528-6