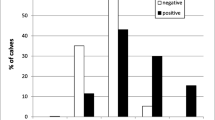
Abstract
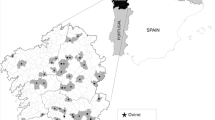
Cryptosporidium parvum is the second-most prevalent Cryptosporidium species that infects humans worldwide. In European countries, it is the most prevalent species in sheep, suggesting that these animals are a source of zoonotic infection. Preweaned lambs and goats are particularly susceptible to infection by the parasite and may suffer from severe diarrhea whilst excreting large quantities of infectious oocysts. Fifty fecal samples from preweaned lambs and goats with diarrhea from 35 farms across Israel, found to be Cryptosporidium-positive by microscopy, were tested by PCR and sequence analyses to determine the infective species and subtypes. Cryptosporidium parvum DNA was detected in most samples from both lambs and goats (46/50). Cryptosporidium xiaoi DNA was detected in three samples from kids, with co-infection detected in a single sample. Eleven different C. parvum subtypes were found, 10 in lambs and 5 in goats. All subtypes were from the IIa and IId subtype families, with subtypes IIdA20G1 and IIaA15G2R1 being the most prevalent and widespread. These subtypes were previously found in calves and humans in Israel and are considered the most prevalent C. parvum subtypes in small ruminants globally. These results underline the zoonotic potential of C. parvum from small ruminants and the high subtype diversity compared to previous reports from other Middle Eastern countries. In addition, this is the first report of C. xiaoi in Israel.

Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F (2003) Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol 41(6):2744–2747
Chalmers RM, Robinson G, Elwin K, Elson R (2019) Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasit Vectors 12(1):95
Chen Y, Qin H, Huang J, Li J, Zhang L (2022) The global prevalence of Cryptosporidium in sheep: a systematic review and meta-analysis. Parasitology 149(12):1652–1665
Fayer R, Santín M (2009) Cryptosporidium xiaoi n. sp. (Apicomplexa: Cryptosporidiidae) in sheep (Ovis aries)) Vet Parasitol 164(2-4):192–200
Feng Y, Li N, Duan L, Xiao L (2009) Cryptosporidium genotype and subtype distribution in raw wastewater in Shanghai, China: evidence for possible unique Cryptosporidium hominis transmission. J Clin Microbiol 47(1):153–157
Grossman T, Ken-Dror S, Pavlotzky E, Vainer J, Glazer Y, Sagi O, Peretz A, Agmon V, Marva E, Valinsky L (2019) Molecular typing of Cryptosporidium in Israel. PLoS One 14(9):e0219977
Guo Y, Li N, Ryan U, Feng Y, Xiao L (2021) Small ruminants and zoonotic cryptosporidiosis. Parasitol Res 120(12):4189–4198
Helmy YA, Hafez HM (2022) Cryptosporidiosis: from prevention to treatment, a narrative review. Microorganisms 10(12):2456
Hijjawi N, Zahedi A, Al-Falah M, Ryan U (2022) A review of the molecular epidemiology of Cryptosporidium spp. and Giardia duodenalis in the Middle East and North Africa (MENA) region. Infect Genet Evol 98:105212
Holzhausen I, Lendner M, Göhring F, Steinhöfel I, Daugschies A (2019) Distribution of Cryptosporidium parvum gp60 subtypes in calf herds of Saxony. Germany. Parasitol Res 118(5):1549–1558
Israeli dairy board. https://www.halavi.org.il. Accessed 14.5.2023
Israeli Ministry of Agriculture (2022) Current status of small ruminant farming in Israel. https://www.gov.il/he/departments/publications/reports/overview-the-sheep-industry-in-israel). Accessed 14.5.2023
Israeli Veterinary Services (2022) Annual report on livestock importation for 2022. https://www.gov.il/he/departments/publications/reports/import_06-2022. Accessed 14.5.2023
Khan A, Shaik JS, Grigg ME (2018) Genomics and molecular epidemiology of Cryptosporidium species. Acta Trop 184:1–14
Markovics A, Elad D, Pipano E (1984) The prevalence of Cryptosporidium in dairy calves in Israel. IJVM 41(4):134–139
Nader JL, Mathers TC, Ward BJ, Pachebat JA, Swain MT, Robinson G, Chalmers RM, Hunter PR, van Oosterhout C, Tyler KM (2019) Evolutionary genomics of anthroponosis in Cryptosporidium. Nat Microbiol 4(5):826–836
R Project for Statistical Computing. https://www.r-project.org/
Ryan U, Zahedi A, Feng Y, Xiao L (2021) An update on zoonotic Cryptosporidium species and genotypes in humans. Animals 11(11):3307
Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, Iqbal J, Khalid N, Xiao L (2005) Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol 43(6):2805–2809
Tanriverdi S, Markovics A, Arslan MO, Itik A, Shkap V, Widmer G (2006) Emergence of distinct genotypes of Cryptosporidium parvum in structured host populations. Appl Environ Microbiol 72:2507–2513
Thompson HP, Dooley JS, Kenny J, McCoy M, Lowery CJ, Moore JE, Xiao L (2007) Genotypes and subtypes of Cryptosporidium spp. in neonatal calves in Northern Ireland. Parasitol Res 100(3):619–624
Thomson S, Innes E, Jonsson N, Katzer F (2016) A multiplex PCR test to identify four common cattle-adapted Cryptosporidium species. Parasitology Open 2:E5
Utaaker KS, Chaudhary S, Kifleyohannes T, Robertson LJ (2021) Global goat! Is the expanding goat population an important reservoir of Cryptosporidium? Front Vet Sci 5(8):648500
Wang T, Guo Y, Roellig DM, Li N, Santín M, Lombard J, Kváč M, Naguib D, Zhang Z, Feng Y, Xiao L (2022) Sympatric recombination in zoonotic Cryptosporidium leads to emergence of populations with modified host preference. Mol Biol Evol 39(7):msac150
World Organization for Animal Health (2022) Cryptosporidiosis – in: Terrestrial Manual, Chapter 3.10.2. https://www.woah.org/en/what-we-do/standards/codes-and-manuals/terrestrial-manual-online-access/. Accessed 14.5.2023
Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, Fayer R, Lal AA (1999) Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol 65(4):1578–1583
Xiao L, Alderisio K, Limor J, Royer M, Lal AA (2000) Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl Environ Microbiol 66(12):5492–5498
Yang R, Jacobson C, Gordon C, Ryan U (2009) Prevalence and molecular characterization of Cryptosporidium and Giardia species in pre-weaned sheep in Australia. Vet Parasitol 161(1-2):19–24
Yasur-Landau D, Zilberberg M, Perry Markovich M, Behar A, Fleiderovitz L, Leszkowicz Mazuz M (2021) Cryptosporidium parvum subtypes from diarrheic dairy calves in Israel. Vet Parasitol Reg Stud Reports 25:100608
Acknowledgements
The study was carried out in part as the final thesis of a DVM student (S. Tako) at the Koret School of Veterinary Medicine, Hebrew University of Jerusalem.
Funding
The study was supported by internal funds from the Kimron Veterinary Institute.
Author information
Authors and Affiliations
Contributions
S.T. and D.Y.L. wrote the manuscript. S.T. and L.F. performed the research. M.P.M. drew the maps. S.T., M.P.M., M.L.M., A.B., and D.Y.L. reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Section Editor: Lihua Xiao
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tako, S., Fleiderovitz, L., Markovich, M.P. et al. Cryptosporidium parvum gp60 subtypes in diarrheic lambs and goat kids from Israel. Parasitol Res 122, 2237–2241 (2023). https://doi.org/10.1007/s00436-023-07925-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-023-07925-0