Abstract
Forensic DNA analysis in compromised skeletal remains may pose challenges due to DNA degradation, often resulting in partial or negative autosomal STRs profiles. To address this issue, alternative approaches such as mitochondrial DNA or SNPs typing may be employed; however, they are labour-intensive and costly. Insertion-null alleles (INNULs), short interspersed nuclear elements, have been suggested as a valuable tool for human identification in challenging samples due to their small amplicon size. A commercial kit including 20 INNULs markers along with amelogenin (InnoTyper® 21) has been developed. This study assesses its utility using degraded skeletal remains, comparing the results obtained (the number of detected alleles, RFU values, PHR, and the number of reportable markers) to those obtained using GlobalFiler™. Subsequently, the random match probability of the two profiles for each sample was determined using Familias version 3 to evaluate the power of discrimination of the results obtained from each kit. In every sample, InnoTyper® 21 yielded more alleles, higher RFU values, and a greater number of reportable loci. However, in most cases, both profiles were similarly informative. In conclusion, InnoTyper® 21 serves as a valuable complement to the analysis of challenging samples in cases where a poor or negative profile was obtained.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Short Tandem Repeats (STRs) represent a well-established method in forensic science for human identity testing applications [1]. However, forensic DNA analysis of degraded skeletal remains can be challenging due to DNA fragmentation and molecular damage resulting from decomposition and environmental exposure [2]. This phenomenon is linked to environmental factors such as temperature [3], humidity [4], salinity, and low pH values [5].
Commercial STR kits typically produce amplicons ranging from 100 to 500 base pairs (bp) [6], potentially resulting in partial or even negative profile due to DNA degradation. To address this issue, smaller STRs, known as miniSTRs, were developed by redesigning primer binding sites to reduce amplicon size [7]. Another approach for dealing with highly degraded sample is mitochondrial DNA (mtDNA) analysis, which involves the use of overlapping small-sized amplicons. However, this method is both labour-intensive and costly [8]. Similar challenges arise with single nucleotide polymorphisms (SNPs) [9].
Insertion and deletion polymorphisms fall somewhere between STRs and SNPs, and they offer the advantage of being compatible with routine capillary electrophoresis based workflows [10]. Retrotransposable elements (Res) include long and short interspersed nuclear elements (LINEs and SINEs respectively). Among SINEs, Alu sequences contain insertion and null alleles (INNULs) that differ in length [11]. A commercial kit comprising 20 INNULs markers, designed with a three-primers strategy (including a common forward primer for both alleles and a specific one for insertion and null alleles) was developed and marketed as InnoTyper® 21 [12].
INNULs typing offers several advantages, including: small amplicon size [13], absence of stutter artifacts [14], and a low mutation rate. However, a significant disadvantage is the substantial difference in length between insertion and null alleles, leading to a preferential amplification of the smaller one [15]. These characteristics make INNULs applicable in human identification [16], the analysis of degraded samples [17], the interpretation of mixtures [18], and population studies and biogeographical ancestry [19].
Our laboratory focuses on identifying the victims of the Spanish Civil War (1936–1939) and the postwar period in Andalusia -southern Spain- under an agreement between the Andalusia local government and our university. The remains we analyse here are highly degraded and often result in partial profiles. As a result, we continually explore alternative approaches to enhance the available information.
The aim of this research is to assess the performance of the InnoTyper® 21 commercial kit for INNULs in a large set of highly degraded skeletal remains samples and compare its efficiency to the autosomal STR kit Globalfiler™ while considering the discriminatory power of both kits.
Material and methods
The following procedures were conducted in a low copy number DNA facility adhering to international standards for ancient DNA work [20,21,22]. This facility is equipped with measures to prevent contamination, including High Efficiency Particle Arresting (HEPA) filtered air positive pressure, a C type ultraviolet room for decontamination, DNAZap™ surfaces decontamination and the use sterile material. In addition to preventive measures, contamination detection protocols were implemented. These protocols involved identifying degraded DNA characteristics such as low quantification results, a high degradation index, and ‘ski-slope’ profiles. Furthermore, profiles of laboratory staff were regularly compared to those obtained from the samples.
Samples
A total of 70 skeletal remains (see Table 1) retrieved from mass graves in Andalusia were analysed in this study. These samples were selected for comparison when a partial or negative Globalfiler™ profile was obtained. The remains had been buried at a depth of 3–4 m for 70–80 years in a region characterized by high temperatures in summer (an average 28ºC with maximums of 45ºC), over 2800 h of solar radiation, low precipitation (400–600 mm rain gauge on rare rainy days) [23], and slightly acidic soil [24].
The surface of the samples was sanded with a Dremel® rotatory tool [25]. Subsequently, the bones were cut into fragments and exposed to UV light for 10 min each side in a 6 W cabin [26]. Tooth and bone fragments were pulverized using a TissueLyser II (QIAGEN, Hilden, Germany). The resulting tooth or bone powder was transferred to a 15-ml Falcon tube.
DNA extraction
DNA from the samples was extracted using an in-house procedure based on the phenol/chloroform/isoamyl alcohol protocol, chosen for its ability to yield higher DNA amounts [27]. One gram of bone or tooth powder was mixed with 5 ml of lysis buffer containing EDTA, proteinase K, SDS (sodium dodecyl sulphate) and DTT (dithiothreitol), and then incubated at 56 ºC overnight. The lysate was mixed with phenol/chloroform/isoamyl alcohol (25:24:1) and the supernatant was concentrated using Amicon® Ultra-4 centrifugal filter unit (Merck, KGaA, Darmstadt, Germany). The extracts were purified using the MinElute® PCR Purification Kit (QIAGEN, Hilden, Germany).
DNA quantification
The purified extracts were quantified by the Quantifiler™ Trio quantitative PCR commercial kit following the manufacturer’s instructions [28]. The qPCR reaction was carried out in a QuantStudio® 5 (ThermoFisher).
DNA amplification
The DNA extracts were amplified using the Globalfiler™ autosomal STR commercial kit (ThermoFisher, Waltham, MA, USA), and the Innotyper® 21 INNULs commercial kit (InnoGenomics, New Orleans, LA) following the manufacturer’s recommendations in both cases [29, 30]. The same amount of DNA extract (15 µl) was added to each reaction.
Data analysis
The amplified samples were subsequently analysed using a 3500 Genetic Analyzer, following the injection parameters provided by each manufacturer. The raw data was analysed using GeneMapper™ IDX v1.6. Four parameters were analysed: 1) the number of detected alleles (alleles above the analytical threshold of 50 RFU for Globalfiler™, and 85 RFU for InnoTyper® 21), 2) average RFU (relative fluorescence units), 3) average PHR (peak height ratio, calculated as the ratio of the smaller allele to the larger allele in heterozygous loci), and 4) the number of reportable loci (markers with homozygous alleles above stochastic threshold, 360 RFU for Globalfiler™ and 180 RFU for InnoTyper® 21, as well as markers with heterozygous alleles above analytical threshold and PHR above 0.60 for Globalfiler™ and 0.76 for InnoTyper® 21). All thresholds were established following internal validation in accordance with SWGDAM’s guidelines [31].
Statistical parameters (mean, coefficient of quartile variation (CQV), Shapiro–Wilk test for normality, Levene’s test for homogeneity of variances, and One-Way ANOVA, including both Welch’s and Fisher’s, were calculated using jamovi 2.2.5 [32].
To evaluate the power of discrimination of each obtained profile, random match probability (RMP) was calculated using Familias version 3 [33]. The calculations utilized 2023 GHEP-ISFG STRs allele frequencies [34] and an earlier publication regarding InnoTyper® 21 allele frequencies in the Andalusian population [19].
Results and discussion
The results of the genetic profiles are summarised in Table 2, presenting both average values and data dispersion as the coefficient of quartile variation. More than 90% of the samples yielded low quantification results for the small DNA target (refer to Supplementary Data), and approximately 85% of the samples yielded five or fewer reportable autosomal STR markers. Within this subset (5 informative markers or less), 32% of them resulted in a negative profile (with no reportable markers). The best Globalfiler™ profile (14 markers) was obtained with a total of 0.738 ng of DNA input, while the best InnoTyper™ result (19 markers) was obtained with a total of 0.091 ng of DNA input, highlighting the higher sensibility of the latter.
In Fig. 1, boxplots depict the analysed variables of genetic profiles. In general terms, InnoTyper® 21 outperforms Globalfiler™ in the number of detected alleles and RFU, reaching approximately twice as many in both cases. Peak height ratios were nearly identical in both kits. Concerning the number of reportable loci, InnoTyper® 21 tripled the count.
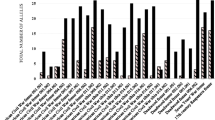
Figure 2 illustrates the number of reportable markers obtained in each sample by kit. Out of the 22 samples that failed to produce reportable markers, 15 of them yielded a positive result with InnoTyper® 21, although they were mostly partial profiles. In many cases, when 5 markers or fewer were obtained by Globalfiler™, InnoTyper® 21 managed to achieve at least half of the profile.
After conducting Shapiro–Wilk and Levene tests, One-Way ANOVA revealed statistically significant differences between the two kits concerning the number of detected alleles (p-value < 0.001), average RFU (p-value = 0.003), and the number of reportable loci (p-value < 0.001). No statistically significant differences were found in terms of peak height ratio (p-value = 0.808).
Previous studies have indicated that InnoTyper® 21 detected more alleles than autosomal STR kits like NGM™ [35] and Globalfiler™ [12, 17, 36], resulting in superior profiles [17, 36], and informative profiles even when Globalfiler™ yielded zero markers [12, 17]. Our findings align with these results, as InnoTyper® 21 obtained more detected alleles and a higher number of reportable markers. However, the results from skeletal remains with prior Gobalfiler™ negative profiles are not as informative as the InnoTyper™ 21 profiles in the study conducted by Martins et al. [17]. These differences may be attributed to their samples being casework rootless hairs.
Nevertheless, there are limited examples in the published literature where not only the number of markers reached by each autosomal STRs/INNULs kit is discussed, but also the statistical significance of both profiles. This evaluation is crucial to determine if InnoTyper® 21 results hold enough power of discrimination for potential reporting.
To assess this, the random match probability (RMP) was calculated using Familias version 3 software for each Globalfiler™/InnoTyper® 21 kit. Figure 3 displays the common logarithm (base 10) of each profile by sample and by commercial kit.
When comparing both Figs. 2 and 3, it is evident that InnoTyper® 21 yields more markers. Nevertheless, its power of discrimination aligns with that already achieved by GlobalFiler™. In fact, no statistically significant differences were observed between the two groups of likelihood ratios (p-value = 0.321) in One-Way ANOVA.
InnoGenomics tested InnoTyper® 21 with sonicated samples and concluded that higher random match probability values were achieved when DNA fragments were < 150 bp compared to the values obtained by Globalfiler™ [37]. Our research supports this idea, as InnoTyper® 21 outperforms Globalfiler™ in terms of random match probability when fewer than 5 markers were achieved with the autosomal STRs commercial kit. However, it is true that the power of discrimination of GlobalFiler™ is astronomical compared to InnoTyper® 21 when 10 or more markers are achieved by the former.
This is why many authors suggests InnoTyper® 21 as a valuable complement to autosomal STRs [12, 35, 38]. Some even propose that the power of discrimination offered by InnoTyper® 21 is higher than the provided by mitochondrial DNA [17]. This is especially true when little markers or a negative profile have been achieved with autosomal STRs approaches.
One last point to consider is whether both likelihood ratios obtained from Globalfiler™ and InnoTyper® 21 can be combined. This topic has been widely discussed in literature, with two main positions: one advocating for the avoidance of combining different DNA evidence [39], and the other one supporting the combination using the product rule [40], with the associated mathematical refinement over time [41]. The key factor is demonstrating the independence between autosomal STRs and INNULs, a question that remains unanswered in the literature. However, a roughly calculated Kosambi recombination fraction [42] gives extremely low values to the markers located in the same loci: AC4027 and D7S820, TARBP and D1S1656, and NBC106 and FGA.
Conclusion
The aim of this research was to evaluate the applicability of InnoTyper® 21 for the analysis of degraded skeletal remains’ DNA and to estimate the power of discrimination obtained from partial INNULs profiles compared to an autosomal STRs approach. A total of 70 degraded skeletal remains samples were typed with Innotyper® 21, specifically selected when a negative or partial profile was obtained by Globalfiler™. InnoTyper® 21 consistently yielded more alleles, higher RFU values, and more reportable loci than Globalfiler™ in every sample. However, despite these differences in profiling, the random match probability values from both profiles were similar. In conclusion, InnoTyper® 21 emerges as a robust complementary tool for addressing partial or negative results in challenging samples.
Data availability
The data supporting the findings of this study are available within the paper and its Supplementary Information.
References
Butler JM (2018) Short tandem repeat typing technologies used in human identity testing. Biotechniques 43:2–5. https://doi.org/10.2144/000112582
Dabney J, Meyer M, Pääbo S (2013) Ancient DNA Damage. Cold Spring Harb Perspect Biol 5:a012567
Al-Kandari NM, Singh J, Sangar VC (2016) Time-dependent effects of temperature and humidity on quantity of DNA in samples of human saliva, blood and semen in Kuwait. Int J Pharm Sci Res 14:2852–2873
Burger J, Hummel S, Herrmann B, Henke W (1999) DNA preservation: A microsatellite-DNA study on ancient skeletal remains. Electrophoresis 20:1722–1728
Creighton TE (2010) The biophysical chemistry of nucleic acids & proteins. Helvetian Press
Butler JM (2007) Short tandem repeat typing technologies used in human identity testing. BioTechniques 43:Sii-Sv. https://doi.org/10.2144/000112582
Butler JM, Shen Y, McCord BR (2003) The Development of Reduced Size STR amplicons as tools for analysis of degraded DNA. J Forensic Sci 48:1054–1064. https://doi.org/10.1520/JFS2003043
Berger C, Parson W (2009) Mini-midi-mito: Adapting the amplification and sequencing strategy of mtDNA to the degradation state of crime scene samples. Forensic Sci Int Genet 3:149–153. https://doi.org/10.1016/j.fsigen.2009.01.011
Pontes ML, Fondevila M, Laréu MV, Medeiros R (2015) SNP Markers as Additional Information to Resolve Complex Kinship Cases. Transfus Med Hemother 42:385–388. https://doi.org/10.1159/000440832
Weber JL, David D, Heil J et al (2002) Human Diallelic Insertion/Deletion Polymorphisms. Am J Hum Genet 71:854–862
Larue BL, Sinha SK, Montgomery AH et al (2012) INNULs: A novel design amplification strategy for retrotransposable elements for studying population variation. Hum Hered 74:27–35. https://doi.org/10.1159/000343050
Brown H, Thompson R, Murphy G et al (2017) Development and validation of a novel multiplexed DNA analysis system, InnoTyper® 21. Forensic Sci Int Genet 29:80–99. https://doi.org/10.1016/j.fsigen.2017.03.017
AnniLauri Villeme, Gretchen Bartizal, Carolyn R. Steffen, Michael D. Coble (2016) Examination of 20 Retrotransposable Polymorphic Insertion/Null (INNUL) markers for their utility in kinship testing using a commercial software program (LSAM). In: Poster B142 – AAFS Annual Meeting Las Vegas, NV, February 26. https://innogenomics.com/wp-content/uploads/2020/11/NIST_AAFS_2016_Poster_Examination_of_20_Retrotransposable_Polymorphic_INNUL_markers_and_LSAM_Final.cleaned.pdf. Accessed 28 Jul 2023
Sudhir K. Sinha, Anne H. Montgomery, Gina Pineda, et al (2018) Development of a novel multiplexed DNA analysis system for highly degraded DNA samples. In: 24th International Symposium on Human Identification. https://www.promega.com/-/media/files/resources/conference-proceedings/ishi-24/oral-presentations/sinha-abstract.pdf. Accessed 28 Jul 2023
Peters D (2016) Use of InnoTyper21 with Highly Degraded Missing Persons Samples. https://innogenomics.com/wp-content/uploads/2020/11/Peters_2016_IT21.cleaned.pdf. Accessed 28 Jul 2023
Moura-Neto RS, Mello ICT, Silva R et al (2018) Evaluation of InnoTyper® 21 in a sample of Rio de Janeiro population as an alternative forensic panel. Int J Legal Med 132:149–151. https://doi.org/10.1007/s00414-017-1642-2
Martins C, Ferreira PM, Carvalho R et al (2019) Evaluation of InnoQuant® HY and InnoTyper® 21 kits in the DNA analysis of rootless hair samples. Forensic Sci Int Genet 39:61–65. https://doi.org/10.1016/j.fsigen.2018.12.005
Kim GH, Kwon SY, Lee S-Y et al (2020) Evaluation of the InnoTyper21® system for the applications into trace and degraded DNA in the Korean population. Int J Legal Med 134:2053–2059. https://doi.org/10.1007/s00414-020-02429-3
Saiz M, Gutierrez-Carvajal A, Galvez X et al (2019) Population genetic data of 20 insertion-null allele markers in the Andalusian population. Int J Legal Med 133:1397–1400. https://doi.org/10.1007/s00414-019-02044-x
Cooper A, Poinar H (2000) Ancient DNA: Do It Right or Not at All. Science 289:1139
Green RE, Briggs AW, Krause J et al (2009) The Neandertal genome and ancient DNA authenticity. EMBO J 28:2494–2502. https://doi.org/10.1038/emboj.2009.222
Pääbo S, Poinar H, Serre D et al (2004) Genetic Analyses from Ancient DNA. Annu Rev Genet 38:645–679. https://doi.org/10.1146/annurev.genet.37.110801.143214
Consejería de Sostenibilidad, Medio Ambiente y Economía Azul de la Junta de Andalucía (2022) Caracterización media de las principales variables climáticas en Andalucía. https://www.juntadeandalucia.es/medioambiente/portal/areas-tematicas/cambio-climatico-y-clima/clima-en-andalucia/caracterizacion_media_principales_variables_climaticas_andalucia. Accessed 28 Jul 2023
Consejería de Medio Ambiente de la Junta de Andalucía (1999) “Los criterios y estándares para declarar un suelo contaminado en Andalucía y la metodología y técnicas de toma de muestra y análisis para su investigación.” https://www.ugr.es/~fjmartin/INFORMES/Criterios%20y%20estandares.pdf. Accessed 28 Jul 2023
Marjanović D, Durmić-Pašić A, Bakal N et al (2007) DNA Identification of Skeletal Remains from World War II Mass Graves Uncovered in Slovenia Forensic Science Forensic Science. Croat Med J 48:513–519
Carlyle SW, Parr RL, Hayes MG, O’Rourke DH (2000) Context of Maternal Lineages in the Greater Southwest. Am J Phys Anthropol 113:85–101
Haarkötter C, Gálvez X, Vinueza-Espinosa DC et al (2023) A comparison of five DNA extraction methods from degraded human skeletal remains. Forensic Sci Int 348:111730. https://doi.org/10.1016/j.forsciint.2023.111730
Thermo Fisher Scientific (2018) QuantifilerTM HP and Trio DNA Quantification Kits. In: User Guide. https://assets.thermofisher.com/TFS-Assets/LSG/manuals/4485354.pdf. Accessed 29 Jul 2021
ThermoFisher Scientific (2019) GlobalFilerTM and GlobalFilerTM IQC PCR Amplification Kits. In: User Guide. https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2F4477604.pdf. Accessed 28 Jul 2023
InnoGenomics (2018) InnoTyper® 21 Human DNA Analysis Kit – User Guide v3.8. In: User Guide. https://innogenomics.com/wp-content/uploads/2023/01/InnoTyper_21_User_Guide_v3.8.pdf. Accessed 28 Jul 2023
Scientific Working Group on DNA Analysis Methods (2016) SWGDAM Validation Guidelines for DNA Analysis Methods. https://www.swgdam.org/_files/ugd/4344b0_813b241e8944497e99b9c45b163b76bd.pdf. Accessed 28 Jul 2023
The jamovi project (2024) jamovi (Version 2.5) [Computer Software]. Retrieved from https://www.jamovi.org. Accessed 28 Jul 2023
Kling D, Tillmar AO, Egeland T (2014) Familias 3 – Extensions and new functionality. Forensic Sci Int Genet 13:121–127. https://doi.org/10.1016/j.fsigen.2014.07.004
GHEP-ISFG (Grupo de Habla Española y Portuguesa de la International Society for Forensic Genetics) (2022) Tabla de frecuencias españolas alélicas para STRs autosómicos/Autosomal STRs Spanish allele frequencies. https://ghep-isfg.org/usercontent/blobs/2022/03/01/YgsROqxbRNuAaeDU9WQhfg_frequencies_2022.pdf. Accessed 28 Jul 2023
Van Den Berge M, Wiskerke D, Gerretsen RRR et al (2016) DNA and RNA profiling of excavated human remains with varying postmortem intervals. Int J Legal Med 130:1471–1480. https://doi.org/10.1007/s00414-016-1438-9
Martins C, Ferreira PM, Carvalho R et al (2018) Internal validation of two new retrotransposons-based kits (InnoQuant® HY and InnoTyper® 21) at a forensic lab. Forensic Sci Int 283:1–8. https://doi.org/10.1016/j.forsciint.2017.11.037
Brown H, Murphy GP, Montgomery AH, et al (2016) Developmental validation of InnoTyper® 21, a nuclear DNA typing system based on retrotransposable element polymorphisms for degraded forensic samples. In: International Symposium on Human Identification (ISHI). https://innogenomics.com/wp-content/uploads/2021/01/Innogenomics_ISHI_2016_IT21_Dev_Valid_Poster_Final.cleaned.pdf. Accessed 28 Jul 2023
Ristow PG, Barnes N, Murphy GP et al (2017) Evaluation of the InnoTyper® 21 genotyping kit in multi-ethnic populations. Forensic Sci Int Genet 30:43–50. https://doi.org/10.1016/j.fsigen.2017.06.002
Amorim A (2008) A cautionary note on the evaluation of genetic evidence from uniparentally transmitted markers. Forensic Sci Int Genet 2:376–378. https://doi.org/10.1016/j.fsigen.2008.04.001
Walsh B, Redd AJ, Hammer MF (2008) Joint match probabilities for Y chromosomal and autosomal markers. Forensic Sci Int 174:234–238. https://doi.org/10.1016/j.forsciint.2007.03.014
Buckleton J, Myers S (2014) Combining autosomal and Y chromosome match probabilities using coalescent theory. Forensic Sci Int Genet 11:52–55. https://doi.org/10.1016/j.fsigen.2014.02.009
Fondevila M, Santos C, Phillips C, et al (2011) An assessment of linkage between forensic markers: core STRs, mini-STRs and InDels. In: 22nd International Symposium on Human Identification. https://strbase-archive.nist.gov/pub_pres/ISHI2011_Fondevila.pdf. Accessed 28 Jul 2023
Funding
Funding for open access publishing: Universidad de Granada/CBUA. C.H. acknowledges the Spanish Ministry of Universities for funding the development of his PhD (FPU 20/01967).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Informed consent
Not applicable.
Conflict of interest
The authors declare no conflict of interest.
Research involving human participants and/or animals
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Haarkötter, C., Saiz, M., Gálvez, X. et al. Evaluation of the usefulness of insertion-null markers in critical skeletal remains. Int J Legal Med (2024). https://doi.org/10.1007/s00414-024-03205-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00414-024-03205-3