Abstract
Respiratory tract infection (RTI) remains a significant cause of morbidity and mortality across the globe. The optimal management of RTI relies upon timely pathogen identification via evaluation of respiratory samples, a process which utilises traditional culture-based methods to identify offending microorganisms. This process can be slow and often prolongs the use of broad-spectrum antimicrobial therapy, whilst also delaying the introduction of targeted therapy as a result. Nanopore sequencing (NPS) of respiratory samples has recently emerged as a potential diagnostic tool in RTI. NPS can identify pathogens and antimicrobial resistance profiles with greater speed and efficiency than traditional sputum culture-based methods. Increased speed to pathogen identification can improve antimicrobial stewardship by reducing the use of broad-spectrum antibiotic therapy, as well as improving overall clinical outcomes. This new technology is becoming more affordable and accessible, with some NPS platforms requiring minimal sample preparation and laboratory infrastructure. However, questions regarding clinical utility and how best to implement NPS technology within RTI diagnostic pathways remain unanswered. In this review, we introduce NPS as a technology and as a diagnostic tool in RTI in various settings, before discussing the advantages and limitations of NPS, and finally what the future might hold for NPS platforms in RTI diagnostics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Respiratory tract infection (RTI) is a leading cause of mortality globally, as well as a significant cause of morbidity amongst previously healthy adults [1]. Effective treatment of RTI relies upon efficient diagnosis and rapid delivery of appropriate treatment, usually in the form of antimicrobial therapy [2]. Whilst polymerase chain reaction (PCR) technologies have become mainstream within viral RTI diagnostic panels, a practice highlighted by the coronavirus pandemic, their use within bacterial RTI is extremely limited. Diagnostic strategies within this area still rely heavily on traditional culture methods, which are often slow and sometimes generate ambiguous mixed growth results or discard important results as oral flora [3].
Reducing time to pathogen identification in bacterial RTI represents an important focus of study for two reasons. Firstly, delivering a targeted antibiotic sooner would almost certainly improve clinical outcomes, and secondly, it would greatly reduce the use of broad-spectrum antibiotics, attenuating drivers of antimicrobial resistance. Whilst multiplex bacterial PCR improves time to diagnosis in RTI [4], it is third-generation sequencing (TGS) techniques such as Nanopore sequencing (NPS) that offer real promise as a new diagnostic tool.
Nanopore technology can sequence metagenomic samples quickly, requiring basic preparation to generate accurate genomic data that permit pathogen identification [5, 6]. Multiple studies using Nanopore technology to sequence respiratory samples and identify pathogens [7] have demonstrated that NPS is a feasible option as a diagnostic tool in RTI. In this review, we describe the origins of NPS through to its present use and potential future use as a clinical tool in RTI.
Nanopore Sequencing
Origins & Development
NPS was first described in the 1980s with translocation of single-stranded polynucleotides via an electrically charged α-haemolysin (αHL) pore [8]. Associated studies demonstrated the potential to characterise base pairs by monitoring bases’ differing disruption to ionic current. This led to a patent in 1995 by Church, Deamer, Branton and colleagues [9]. Kasianowicz and colleagues worked to improve understanding and throughput of the Nanopore approach by preventing spontaneous pore gating (pore closure) and by demonstrating the capability to sequence both RNA and DNA directly, with indiscriminate initiation from 3′ and 5′ ends [8].
Identification of individual bases was demonstrated in 2005, starting with adenine, and a modified αHL was subsequently shown to be capable of distinguishing between the four bases of DNA within homopolymers and heteropolymers [10, 11]. These approaches were limited by the reliance on DNA immobilisation within the pore. This was later overcome using an MspA mutant pore, which provided higher accuracy due to greater field disruption [12].
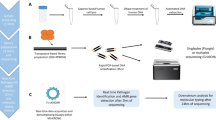
Oxford Nanopore Technologies (ONT) licenced NPS in 2008. This led to the release in 2012 of the MinION, a USB-powered 100 g device capable of rapid, high sensitivity sequencing across 2,048 separately embedded pores. The MinION and other ONT sequencing instruments are controlled and complemented by ever advancing software produced by ONT and third parties. In recent years, packages such as EPI2ME, which offers all-in-one workflow management, have simplified the computational side of the workflow (see Fig. 1). Other key advances in NPS technology in the last decade include improvement in read identity/accuracy from 60–85% to > 99.9%, partially assisted by post-sequencing computational analyses, like consensus calling (Fig. 1) [13, 14], and detection of DNA methylation [15]. Due to its fundamental role in cancer, methylation detection offers a unique diagnostics opportunity [16]. These and other developments are allied with the sensitivity to detect mutants in a 1:100 dilution, as demonstrated in oncological biosensing [17].
Simplified workflow for Nanopore sequencing. A Samples with varying diversity of constituents can be used. B Various isolation protocols can be used, depending on the nature of the desired polynucleotides. C The desired genetic material is often refined along with unwanted genetic material, which in a clinical context would be host DNA. Such unwanted genetic material reduces the concentration of the targeted genetic material, weakening the desired signal during sequencing. Depletion protocols are therefore often used to reduce host DNA, enriching microbial signal. D During library preparation, adapters tagged with motor protein are ligated onto polynucleotides. E Sequencing is initiated by the adapter being guided to an available pore by attachment to a tether. The motor protein then attaches to the pore, where it enzymatically separates the strands into template and complementary strands. The potential difference across the membrane pulls the template strand through the pore, most often releasing the complementary strand, although in 10–20% of cases, the complementary strand is also pulled through the pore to produce a duplex read of both strands. E As the polynucleotide passes through the pore, there is a disruption in current flow which is unique to each of the four bases, and indeed to modified versions of those bases such as 5-methylcytosine. The output of this disruption is the raw signal, which is converted to the sequence of bases in a process known as base calling F. G Some workflows utilise adaptive sampling in which the sequencing output is compared in real time with pre-selected sequences; any strand not matching the pre-selected sequences is ejected from the pore. Adaptive sampling allows enrichment of desired signal, for example signal from microbial DNA versus signal from host DNA. H The resulting reads are aligned, and once sufficient reads have been aligned, high confidence consensus sequences can be formed
NPS Versus Other Third-Generation Sequencing Techniques
TGS methods can produce long reads, facilitating genome sequence assembly and allowing sequencing of complete transcripts. Two TGS modalities exist: single-molecule-real-time (SMRT) sequencing from Pacific Biosciences (PacBio), and NPS from ONT, each including several devices (Table 1) and associated bioinformatics tools. MinION (ONT) is the cheapest TGS device; it is portable and has relatively low infrastructure requirements (Table 1). Unlike previous sequencing methods, particularly those based on PCR which require some level of insight for primer selection/design, TGS methods can be agnostic; by avoiding amplification, TGS methods can produce results which accurately reflect the diversity and proportions of microbial populations in a given sample [4, 18]. This leaves the challenge of differentiating between microbiota and pathogen.
ONT and PacBio platforms are capable of RNA sequencing, typically requiring indirect sequencing via translation to cDNA, often with an amplification step [19]. Nanopore devices are, uniquely, also capable of direct RNA sequencing, providing a theoretical advantage in clinical diagnostics where rapid detection is often vital. In the respiratory tract, for example, where viral disease is prominent, several common viruses are RNA-based, including SARS-CoV-2, respiratory syncytial virus, and influenza A.
Nanopore Sequencing in RTI
The implementation of metagenomics in clinical settings was initially hindered by capital and maintenance costs, requirement for highly skilled staff and uncompetitive turnaround times compared to traditional culture-based methods. More recent sequencing techniques, however, offer much reduced turnaround times, reduced resource and skill requirements, and lower capital and maintenance costs. Nanopore-based sequencing platforms are already being investigated as diagnostic tools in RTI, with promising results reported in a range of clinical settings [7].
Viral Pathogens
Viral pathogens are common drivers of acute RTI in both adults and children. PCR-based methods are already commonplace in diagnostic panels; examples include Influenza A and SARS-CoV-2 diagnostics, where PCR allows rapid and accurate detection of infection [23, 24]. NPS methods have been shown to be as accurate as PCR-based methods with regard to common viral RTI, with the crucial advantage of generating real-time data whilst also being more portable and requiring less laboratory infrastructure [25, 26]. NPS methods, moreover, permit whole viral genome sequencing which enables large-scale epidemiological surveillance, crucial in viruses that have the potential to mutate rapidly in key genomic locations [27].
Bacterial Pathogens
Historically, bacterial RTI diagnostics in clinical settings have been limited, and they continue to be limited, to traditional culture methods, utilising broncho-alveolar lavage (BAL) or sputum samples as substrate [28]. UK guidelines advise a strategy of commencing broad-spectrum antibiotic therapy in patients with RTI, adjusting to more focused therapy once culture results are available [29]. On average, culture results take 48–72 h to be made available to physicians. As a result, patients remain on broad-spectrum antibiotics for extended periods, potentially contributing to the development of antimicrobial resistance. Likewise, the delay in targeted antibiotic therapy negatively affects patient outcomes [30, 31].
NPS has been shown to be an accurate and cost-effective method of diagnosing bacterial RTIs in various clinical settings [7, 32, 33]. Notably, a turnaround time of under 6 h (sample received to pathogen identification time) has been reported for the diagnosis of bacterial RTI [34], including data on antibiotic resistance profiles [35]. Such speed would allow patients to receive targeted therapy sooner and would permit sharply reduced use of broad-spectrum antibiotics, leading to better patient outcomes and improved antimicrobial stewardship.
Mycobacterial Pathogens & Pulmonary Tuberculosis
Pulmonary tuberculosis represents a huge burden on global respiratory health and is often complicated by patterns of multi-drug resistance [36, 37]. PCR-based diagnostic methods are in use for suspected cases of pulmonary tuberculosis [38], but traditional culture of BAL or sputum sample remains the gold standard for diagnosis [39]. Culture results can take 4–6 weeks to be confirmed, representing a significant area of diagnostic delay [40]. Non-tuberculous mycobacteria pulmonary disease (NTM-PD), moreover, is rising in incidence globally [41]. NTM-PD is driven by a multitude of mycobacterial species, and is again diagnosed by traditional mycobacterial culture, which often takes 4–6 weeks to process and obtain results [42]. PCR-based methods are less common here and are often targeted at Mycobacterium tuberculosis rather than mycobacterial species more broadly [43]. NTM-PD therapy may therefore be delayed until confirmatory culture results are received, with potential negative effects on patient outcomes. NPS is effective in detecting mycobacterial species (including Mycobacterium tuberculosis) in respiratory samples, with accuracy and turnaround times comparable to those mentioned above for bacterial RTI [44, 45].
Advantages and Limitations of Nanopore Sequencing
Read Length
NPS can output ultra-long reads, as previously emphasised. For example, consistent read lengths on the order of 880 kb have been reported in non-clinical specimens [15]. Ultra-long reads can, however, run into error rate issues when translocation speed slows in the later stages of runs. This issue can now be resolved using’refuelling buffer’ [46], reflecting ongoing innovation that has made NPS more reliable and capable, regardless of target sequence, qualities that are vital for robust diagnostic tools. Clinically, long read lengths can allow for whole-genome sequencing of smaller (mainly viral) genomes and thus potentially produce highly specific diagnostic tests and outbreak surveillance platforms. Moreover, long read lengths can potentially aid in the investigation of antimicrobial resistance by sequencing (in a single read) complex areas of pathogen genomes where these genes may be found.
Time to Antimicrobial Resistance Data
Treatment efficacy in infectious disease relies heavily on the speed with which appropriate treatment is identified. Antibiotic Susceptibility Tests (ASTs) are currently the gold standard as they identify phenotypical resistance. A proof-of-principle study conducted on Klebsiella pneumoniae, testing NPS’s ability to replace current ASTs, found 80–100% concurrence (averaging 92%) between ASTs and NPS results, when NPS is paired with computational assembly. Whilst ASTs can take > 72 h, results from NPS were available after 38 h [47]. NPS time to result can be reduced using more specialised approaches such as saponin-based depletion of host DNA. In the case of E. coli, for example, the resistance genes blaTEM, sulf1 and dfrA17 were undetected after two hours of sequencing untreated samples, besides one alignment of sulf1. In saponin-depleted samples, in contrast, all three of these resistance genes were detected within the first 20 min [7]. Similar improvements can be achieved via other methods. An enzyme-based host depletion protocol in combination with adaptive sequencing (Fig. 1), for example, showed a 113.41-fold increase in the median of microbial reads [48]. These results demonstrate that NPS, when paired with efficient host DNA depletion, is capable of rapid and broad AMR detection. The primary constraint is currently translation of genotype into phenotypical resistance.
Accuracy & Sensitivity
NPS is sometimes criticised for relatively low raw output accuracy, with base calling errors previously encompassing 5–25% of a given sequence [8]. Whilst NPS accuracy has been relatively low amongst TGS methods, it has improved and is still improving through changes to the pore protein and sequencing chemistry, alongside developments in software. In a recent iteration, ONT claimed 99.6% raw accuracy using flow cell R10.4.1 and ‘‘super accuracy’’ base calling [49]. In practice, R9.4.1 achieved 95.5–98% accuracy in a diverse-activated sludge microbiome, when paired with Guppy V6.0.0 base-caller [50]. NPS paired with consensus calling has achieved 100% accuracy for SARS-CoV-2 when compared with Sanger sequencing, the current gold standard for clinical research [51].
NPS has already been shown in some cases to be more sensitive than traditional diagnostics, and the benefits of agnostic metagenomics in identification of culture-negative pathogens have been highlighted [52]. More recently, the sensitivity and specificity of NPS diagnoses, assessed using patient samples, were found to be 94.5% and 31.8%, respectively, across fungal and bacterial infections; NPS was found to be 56.7% more accurate with regard to true positives than culture methods, whilst retaining inaccuracy in true negatives [53]. For viral pathogens, the sensitivity and specificity are much higher, with 99.1 and 99.6%, respectively, for SARS-CoV-2 [54], and 100% sensitivity for avian Influenza A [55].
Combining NPS With Other Methods
As previously mentioned, NPS has several unique benefits. In addition, numerous methods exist to enhance its suitability for clinical use, though these can introduce other issues. Using a Cas9/sgRNA complex to protect desired sequences from exonucleases, for example, provides an effective ‘selective enrichment’ alternative to depletion protocols. This approach is hindered, however, by a restriction to short reads and a requirement for knowledge of the target(s) [56].
Using the MinION to sequence amplicons produced by a PCR assay has provided high confidence positive results with bacterial pathogens within the first ten minutes of sequencing. This involved a diagnostic pipeline capable of processing 45 samples across 12 h; though this was hindered, however, by excessive false positives/negatives [57]. Combining high-throughput NGS platforms and NPS has found success in pathogen and AMR detection; this method took 212 h (from sample to results), 12 h of which were the NPS stage [58]. Such methods have shown greater precision and accuracy than culture methods but at the cost of a considerably longer time requirement with consequent increased demand for resources and skilled staff, compared to methods using only NPS.
Sample Requirements & Analysis
An important limitation of NPS is the requirement for relatively large amounts of sample, currently up to a few micrograms of DNA and hundreds of nanograms of RNA. Considering that biomedical investigations often rely on a limited amount of genetic material, reducing the amount of sample needed for NPS would promote uptake. More user-friendly bioinformatics platforms and sufficient cloud storage would further promote the application of NPS in clinical settings.
Additional Diagnostic Roles
Outbreak Surveillance
NPS can be used for real-time and field genomic surveillance of potential new infectious diseases. These epidemiological and phylogenetic investigations can result in the timely identification of potential diagnostic targets and treatments, as well as monitoring the evolution and transmission rate of the new infection. For example, NPS was used to conduct genomic surveillance of the yellow fever virus [59], Zika virus [60] and dengue virus [61] worldwide. A Salmonella outbreak in an American hospital was identified using NPS with all positive cases reported within 2 hours [62]. Real-time genomic surveillance using NPS was carried out in Guinea for the ongoing Ebola virus outbreak [63]. Real-time genomic surveillance has been more recently applied to pathogens with large genomes including bacteria such as K. pneumoniae and N. meningitidis [64, 65], fungal pathogens such as Candida auris [66], and large viruses such as Lassa fever virus [67], Zika virus [60], Venezuelan equine encephalitis [68] and SARS-CoV-2 coronavirus [51].
Other Infectious Diseases
Thanks to its real-time sequencing capability, NPS has been used for rapid pathogen detection in prosthetic joints [69], bacterial meningitis [70], infective endocarditis [52] and pneumonia [71]. In six retrospective meningitis cases, NPS detected pathogenic bacteria in only ten minutes, corroborating the idea that NPS can permit early administration of antibiotics following the timely identification of pathogenic bacteria [70]. It is worth mentioning that NPS can additionally be used for the investigation of antimicrobial/antibiotic resistance in different microbes. For instance, NPS was used to detect 51 acquired resistance genes from clinical urine samples with no need for culture [5]. More recently, NPS was used to identify resistance genes to colistin in 12,052 strains of Salmonella [72]. Thanks to the ability to perform longer reads, NPS is a robust technology for the identification of virulent strains and species, ultimately providing a reliable estimate of microbiome composition [73, 74].
Conclusion
NPS has advanced significantly as a diagnostic platform over the last decade and has now reached the tipping point of becoming a feasible addition to clinical diagnostic panels within hospitals and medical outreach centres. The advantages of NPS are multitude and provide clear clinical benefit (reduced time to pathogen identification, broader search panels), as well as public health benefits with regards to reducing inappropriate antimicrobial use and improved outbreak surveillance. The associated cost, training and infrastructure required to establish NPS platforms have, to date, limited its widespread adoption, but these aspects continue to improve and are the focus of ongoing research within the field. With this in mind, NPS technology should be initially instituted in clinical areas where it can be most effective and confers the largest benefit to patients. RTI is an obvious target due to the accessibility of testable specimens, large clinical burden and high transmissibility of disease. Further translational research and large-scale trials are needed to test the utility of NPS in clinical microbiology laboratories.
References
Troeger C, Blacker B, Khalil, et al (2018) Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: a systematic analysis for the Global burden of disease study 2016. Lancet Infect Dis 18(11):1191–1210. https://doi.org/10.1016/S1473-3099(18)30310-4
Woodhead M, Blasi F, Ewig S, Guidelines for the management of adult lower respiratory tract infections - Full version et al (2011) Guidelines for the management of adult lower respiratory tract infections - Full version. Clin Microbiol Infect 17(s6):E1–E59. https://doi.org/10.1111/j.1469-0691.2011.03672.x
Loeffelholz M, Chonmaitree T (2010) Advances in diagnosis of respiratory virus infections. Int J Microbiol. https://doi.org/10.1155/2010/126049
Darie AM, Khanna N, Jahn K et al (2022) Fast multiplex bacterial PCR of bronchoalveolar lavage for antibiotic stewardship in hospitalised patients with pneumonia at risk of Gram-negative bacterial infection (Flagship II): a multicentre, randomised controlled trial. Lancet Respir Med 10(9):877–887. https://doi.org/10.1016/S2213-2600(22)00086-8
Schmidt K, Mwaigwisya S, Crossman LC et al (2017) Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother 72(1):104–114. https://doi.org/10.1093/jac/dkw397
Wang C, Huang Z, Fang W et al (2020) Preliminary assessment of nanopore-based metagenomic sequencing for the diagnosis of prosthetic joint infection. Int J Infect Dis 97:54–59. https://doi.org/10.1016/j.ijid.2020.05.044
Charalampous T, Kay GL, Richardson H et al (2019) Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol 37(7):783–792. https://doi.org/10.1038/s41587-019-0156-5
Deamer D, Akeson M, Branton D (2016) Three decades of nanopore sequencing. Nat Biotechnol 34(5):518–524. https://doi.org/10.1038/nbt.3423
Church, G., Deamer, D.W., Branton, D., et al., 2013. Characterization of individual polymer molecules based on monomer-interface interactions [Online]. EP1956367B1. Available from: https://patents.google.com/patent/EP1956367B1/en?q=Church+Deamer+Branton+nanopore&before=priority:19951231&after=priority:19950101&oq=Church+Deamer+Branton+nanopore+1995 [Accessed 9 Jan 2023]
Ashkenasy N, Sánchez-Quesada J, Bayley H et al (2005) Recognizing a single base in an individual DNA strand: a step toward DNA sequencing in nanopores. Angewandte Chemie International Edition 44(9):1401–1404. https://doi.org/10.1002/anie.200462114
Stoddart D, Heron AJ, Mikhailova E et al (2009) Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc Nat Academy Sci 106(19):7702–7707. https://doi.org/10.1073/pnas.0901054106
Derrington IM, Butler TZ, Collins MD et al (2010) Nanopore DNA sequencing with MspA. Proc Nat Academy Sci 107(37):16060–16065. https://doi.org/10.1073/pnas.1001831107
Jain M, Fiddes IT, Miga KH et al (2015) Improved data analysis for the MinION nanopore sequencer. Nat Methods 12(4):351–356. https://doi.org/10.1038/nmeth.3290
Lemon JK, Khil PP, Frank KM et al (2017) Rapid Nanopore Sequencing of plasmids and resistance gene detection in clinical isolates. J Clin Microbiol 55(12):3530–3543. https://doi.org/10.1128/JCM.01069-17
Jain M, Koren S, Miga KH et al (2018) Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol 36(4):338–345. https://doi.org/10.1038/nbt.4060
Zhang J, Xie S, Xu J et al (2021) Cancer biomarkers discovery of methylation modification with direct high-throughput nanopore sequencing. Front Genetics. https://doi.org/10.3389/fgene.2021.672804
Norris AL, Workman RE, Fan Y et al (2016) Nanopore sequencing detects structural variants in cancer. Cancer Biol Ther 17(3):246–253. https://doi.org/10.1080/15384047.2016.1139236
Xu Y, Lewandowski K, Downs LO et al (2021) Nanopore metagenomic sequencing of influenza virus directly from respiratory samples: diagnosis, drug resistance and nosocomial transmission, United Kingdom, 2018/19 influenza season. Eurosurveillance 26(27):2000004. https://doi.org/10.2807/1560-7917.ES.2021.26.27.2000004
Depledge DP, Mohr I, Wilson AC (2018) Going the distance: optimizing RNA-seq strategies for transcriptomic analysis of complex viral genomes. J Virol 93(1):e01342-18. https://doi.org/10.1128/JVI.01342-18
Pacific Biosciences, 2022. PacBio Announces Record Orders, Including Orders for 76 Revio Systems Received in the Fourth Quarter of 2022. PacBio [Online]. Available from: https://www.pacb.com/press_releases/pacbio-announces-record-orders-including-orders-for-76-revio-systems-received-in-the-fourth-quarter-of-2022/ [Accessed 16 Jan 2023]
Pacific Biosciences, 2023. Sequencing systems - PacBio [Online]. Available from: https://www.pacb.com/sequencing-systems/ [Accessed 18 Jan 2023]
Oxford Nanopore Technologies, 2023. Products. Oxford Nanopore Technologies [Online]. Available from: https://nanoporetech.com/products [Accessed 18 Jan 2023]
Elf S, Auvinen P, Jahn L et al (2018) Development and evaluation of a rapid nucleic acid amplification method to detect influenza A and B viruses in human respiratory specimens. Diagn Microbiol Infect Dis 92(1):37–42. https://doi.org/10.1016/j.diagmicrobio.2018.04.006
Böger B, Fachi MM, Vilhena RO et al (2021) Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. American J Infect Control 49(1):21–29. https://doi.org/10.1016/j.ajic.2020.07.011
Lewandowski K, Xu Y, Pullan ST et al (2019) Metagenomic nanopore sequencing of influenza virus direct from clinical respiratory samples. J Clin Microbiol 58(1):e00963-19. https://doi.org/10.1128/JCM.00963-19
Li X, Li C, Du P et al (2022) Rapid and accurate detection of SARS coronavirus 2 by nanopore amplicon sequencing. Front Microbiol 13:735363. https://doi.org/10.3389/fmicb.2022.735363
Yakovleva A, Kovalenko G, Redlinger M et al (2021) Tracking SARS-COV-2 variants using nanopore sequencing in Ukraine in Summer 2021. Res square. https://doi.org/10.21203/rs.3.rs-1044446/v1
Loens K, Van Heirstraeten L, Malhotra-Kumar S et al (2009) Optimal sampling sites and methods for detection of pathogens possibly causing community-acquired lower respiratory tract infections. J Clin Microbiol 47(1):21–31. https://doi.org/10.1128/JCM.02037-08
NICE, U.K., 2021. Clinical Knowledge Summary - Community-acquired pneumonia, NICE [Online]. [Online]. Available from: https://cks.nice.org.uk/topics/chest-infections-adult/management/community-acquired-pneumonia/ [Accessed 15 January 2023].
Kuti EL, Patel AA, Coleman CI (2008) Impact of inappropriate antibiotic therapy on mortality in patients with ventilator-associated pneumonia and blood stream infection: a meta-analysis. J Critical Care 23(1):91–100. https://doi.org/10.1016/j.jcrc.2007.08.007
Kumar Anand, Ellis P et al (2009) Initiation of inappropriate antimicrobial therapy results in a fivefold reduction of survival in human septic shock. CHEST 136(5):1237–1248. https://doi.org/10.1378/chest.09-0087
Yang L, Haidar G, Zia H et al (2019) Metagenomic identification of severe pneumonia pathogens in mechanically-ventilated patients: a feasibility and clinical validity study. Respir Res 20(1):265. https://doi.org/10.1186/s12931-019-1218-4
Zhang H, Wang M, Han X et al (2022) The application of targeted nanopore sequencing for the identification of pathogens and resistance genes in lower respiratory tract infections. Front Microbiol. https://doi.org/10.3389/fmicb.2022.1065159
Wu N, Ranjan P, Tao C et al (2021) Rapid identification of pathogens associated with ventilator-associated pneumonia by nanopore sequencing. Respir Res 22(1):310. https://doi.org/10.1186/s12931-021-01909-3
Charalampous T, Richardson H, Kay G et al (2018) Diagnosis of lower respiratory tract Infections using nanopore sequencing. European Respir J. https://doi.org/10.1183/13993003.congress-2018.PA5308
Floyd K, Glaziou P, Zumla A et al (2018) The global tuberculosis epidemic and progress in care, prevention, and research: an overview in year 3 of the End TB era. Lancet Respir Med 6(4):299–314. https://doi.org/10.1016/S2213-2600(18)30057-2
Pontali E, Visca D, Centis R et al (2018) Multi and extensively drug-resistant pulmonary tuberculosis: advances in diagnosis and management. Current Opinion Pulmonary Med 24(3):244–252. https://doi.org/10.1097/MCP.0000000000000477
Wang HY, Lu JJ, Chang CY et al (2019) Development of a high sensitivity TaqMan-based PCR assay for the specific detection of Mycobacterium tuberculosis complex in both pulmonary and extrapulmonary specimens. Sci Rep 9(1):113. https://doi.org/10.1038/s41598-018-33804-1
Gonzalez-Angulo Y, Wiysonge CS, Geldenhuys H et al (2012) Sputum induction for the diagnosis of pulmonary tuberculosis: a systematic review and meta-analysis. European J clin microbiol infect dis 31(7):1619–1630. https://doi.org/10.1007/s10096-011-1485-6
Pfyffer GE, Wittwer F (2012) Incubation time of mycobacterial cultures: how long is long enough to issue a final negative report to the clinician? J Clin Microbiol 50(12):4188–4189. https://doi.org/10.1128/JCM.02283-12
Kumar K, Loebinger MR (2022) Nontuberculous mycobacterial pulmonary disease: clinical epidemiologic features, risk factors, and diagnosis: the nontuberculous mycobacterial series. CHEST 161(3):637–646. https://doi.org/10.1016/j.chest.2021.10.003
Cowman S, van Ingen J, Griffith DE et al (2019) Non-tuberculous mycobacterial pulmonary disease. European Respir J. https://doi.org/10.1183/13993003.00250-2019
Sarro Y, Dit S, Butzler MA, Sanogo F et al (2021) Development and clinical evaluation of a new multiplex PCR assay for a simultaneous diagnosis of tuberculous and nontuberculous mycobacteria. eBioMedicine. https://doi.org/10.1016/j.ebiom.2021.103527
Hall MB, Rabodoarivelo MS, Koch A et al (2022) Evaluation of Nanopore sequencing for Mycobacterium tuberculosis drug susceptibility testing and outbreak investigation: a genomic analysis. Lancet Microbe. https://doi.org/10.1016/S2666-5247(22)00301-9
Bouso JM, Planet PJ (2019) Complete nontuberculous mycobacteria whole genomes using an optimized DNA extraction protocol for long-read sequencing. BMC Genomics 20(1):793. https://doi.org/10.1186/s12864-019-6134-y
Oxford Nanopore Technologies, 2022. Community. Refuelling a sequencing run with Flush Buffer from EXP-FLP002 [Online]. Available from: https://community.nanoporetech.com/posts/refuelling-a-sequencing-ru [Accessed 8 December 2022]
Tamma PD, Fan Y, Bergman Y et al (2018) Applying rapid whole-genome sequencing to predict phenotypic antimicrobial susceptibility testing results among carbapenem-resistant Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother 63(1):e01923-18. https://doi.org/10.1128/AAC.01923-18
Gan M, Wu B, Yan G et al (2021) Combined nanopore adaptive sequencing and enzyme-based host depletion efficiently enriched microbial sequences and identified missing respiratory pathogens. BMC Genomics 22(1):732. https://doi.org/10.1186/s12864-021-08023-0
Oxford Nanopore Technologies, 2022. Accuracy. Oxford Nanopore Technologies [Online]. Available from: http://nanoporetech.com/accuracy [Accessed 12 December 2022].
Liu L, Yang Y, Deng Y et al (2022) Nanopore long-read-only metagenomics enables complete and high-quality genome reconstruction from mock and complex metagenomes. Microbiome 10(1):209. https://doi.org/10.1186/s40168-022-01415-8
Chan WS, Au CH, Lam HY et al (2020) Evaluation on the use of Nanopore sequencing for direct characterization of coronaviruses from respiratory specimens, and a study on emerging missense mutations in partial RdRP gene of SARS-CoV-2. Virol J 17(1):183. https://doi.org/10.1186/s12985-020-01454-3
Cheng J, Hu H, Kang Y et al (2018) Identification of pathogens in culture-negative infective endocarditis cases by metagenomic analysis. Ann Clin Microbiol Antimicrob 17(1):43. https://doi.org/10.1186/s12941-018-0294-5
Fu Y, Chen Q, Xiong M et al (2022) Clinical performance of nanopore targeted sequencing for diagnosing infectious diseases. Microbiol Spectr 10(2):e00270-22. https://doi.org/10.1128/spectrum.00270-22
Peto L, Rodger G, Carter DP et al (2021) Diagnosis of SARS-CoV-2 infection with LamPORE, a high-throughput platform combining loop-mediated isothermal amplification and nanopore sequencing. J Clin Microbiol 59(6):e03271-20. https://doi.org/10.1128/JCM.03271-20
Crossley BM, Rejmanek D, Baroch J et al (2021) Nanopore sequencing as a rapid tool for identification and pathotyping of avian influenza a viruses. J Vet Diagn Investig 33(2):253–260. https://doi.org/10.1177/1040638720984114
Stevens RC, Steele JL, Glover WR et al (2019) A novel CRISPR/Cas9 associated technology for sequence-specific nucleic acid enrichment. PLOS ONE 14(4):e0215441. https://doi.org/10.1371/journal.pone.0215441
Player R, Verratti K, Staab A et al (2020) Comparison of the performance of an amplicon sequencing assay based on Oxford nanopore technology to real-time PCR assays for detecting bacterial biodefense pathogens. BMC Genomics 21(1):166. https://doi.org/10.1186/s12864-020-6557-5
Wang K, Li P, Lin Y et al (2020) Metagenomic diagnosis for a culture-negative sample from a patient with severe pneumonia by nanopore and next-generation sequencing. Front Cell Infect Microbiol. https://doi.org/10.3389/fcimb.2020.00182
Faria NR, Kraemer MUG, Hill SC et al (2018) Genomic and epidemiological monitoring of yellow fever virus transmission potential. Science (New York, N.Y.) 361(6405):894–899. https://doi.org/10.1126/science.aat7115
Faria NR, Quick J, Claro IM et al (2017) Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 546(7658):406–410. https://doi.org/10.1038/nature22401
de Jesus JG, Dutra KR, da Sales FCS et al (2020) Genomic detection of a virus lineage replacement event of dengue virus serotype 2 in Brazil, 2019. Memorias Do Instituto Oswaldo Cruz 115:e190423. https://doi.org/10.1590/0074-02760190423
Quick J, Ashton P, Calus S et al (2015) Rapid draft sequencing and real-time nanopore sequencing in a hospital outbreak of Salmonella. Gen Biol 16(1):114. https://doi.org/10.1186/s13059-015-0677-2
Quick J, Loman NJ, Duraffour S et al (2016) Real-time, portable genome sequencing for Ebola surveillance. Nature 530(7589):228–232. https://doi.org/10.1038/nature16996
Dong N, Yang X, Zhang R et al (2018) Tracking microevolution events among ST11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerging Microbes Infect 7(1):146. https://doi.org/10.1038/s41426-018-0146-6
Brynildsrud OB, Eldholm V, Bohlin J et al (2018) Acquisition of virulence genes by a carrier strain gave rise to the ongoing epidemics of meningococcal disease in West Africa. Proc Nat Academy Sci United States America 115(21):5510–5515. https://doi.org/10.1073/pnas.1802298115
Rhodes J, Abdolrasouli A, Farrer RA et al (2018) Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect 7(1):43. https://doi.org/10.1038/s41426-018-0045-x
Kafetzopoulou LE, Pullan ST, Lemey P et al (2019) Metagenomic sequencing at the epicenter of the Nigeria 2018 Lassa fever outbreak. Science (New York, NY) 363(6422):74–77. https://doi.org/10.1126/science.aau9343
Russell JA, Campos B, Stone J et al (2018) Unbiased strain-typing of arbovirus directly from mosquitoes using nanopore sequencing: a field-forward biosurveillance protocol. Sci Rep 8(1):5417. https://doi.org/10.1038/s41598-018-23641-7
Sanderson ND, Street TL, Foster D et al (2018) Real-time analysis of nanopore-based metagenomic sequencing from infected orthopaedic devices. BMC genomics 19(1):714. https://doi.org/10.1186/s12864-018-5094-y
Moon J, Kim N, Kim T-J et al (2019) Rapid diagnosis of bacterial meningitis by nanopore 16S amplicon sequencing: a pilot study. Int J med microbiol: IJMM 309(6):151338. https://doi.org/10.1016/j.ijmm.2019.151338
Gorrie CL, Mirceta M, Wick RR et al (2018) Antimicrobial-resistant Klebsiella pneumoniae carriage and infection in specialized geriatric care wards linked to acquisition in the referring hospital. Clin Infect Dis 67(2):161–170. https://doi.org/10.1093/cid/ciy027
Lu X, Zeng M, Xu J et al (2019) Epidemiologic and genomic insights on mcr-1-harbouring Salmonella from diarrhoeal outpatients in Shanghai, China, 2006–2016. EBioMedicine 42:133–144. https://doi.org/10.1016/j.ebiom.2019.03.006
Nicholls SM, Quick JC, Tang S et al (2019) Ultra-deep, long-read nanopore sequencing of mock microbial community standards. GigaScience 8(5):giz043. https://doi.org/10.1093/gigascience/giz043
Hu Y, Fang L, Nicholson C et al (2020) Implications of error-prone long-read whole-genome shotgun sequencing on characterizing reference microbiomes. iScience 23(6):101223. https://doi.org/10.1016/j.isci.2020.101223
Funding
The authors have not disclosed any funding.
Author information
Authors and Affiliations
Contributions
RC, LJ, ADA produced the first draft of the manuscript having performed an extensive literature review MA, AS, SB reviewed the manuscript and literature and provided critical revisions of the manuscript All authors reviewed the final manuscript prior to submission
Corresponding author
Ethics declarations
Conflict of interest
RC, AS, MA, SB: report no conflicts of interest or funding related to this work. LJ is a PhD candidate partly funded by Oxford Nanopore Technologies.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chapman, R., Jones, L., D’Angelo, A. et al. Nanopore-Based Metagenomic Sequencing in Respiratory Tract Infection: A Developing Diagnostic Platform. Lung 201, 171–179 (2023). https://doi.org/10.1007/s00408-023-00612-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00408-023-00612-y